Application of ClCAC gene and ClSAND gene as reference genes in analysis of gene expression of watermelon fruits
A gene expression and internal reference gene technology, applied in the field of plant molecular biology, can solve the problems of lack of internal reference genes, affecting the accuracy of quantitative analysis of gene expression, unreasonable internal reference genes, etc., and achieving the effect of wide applicability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
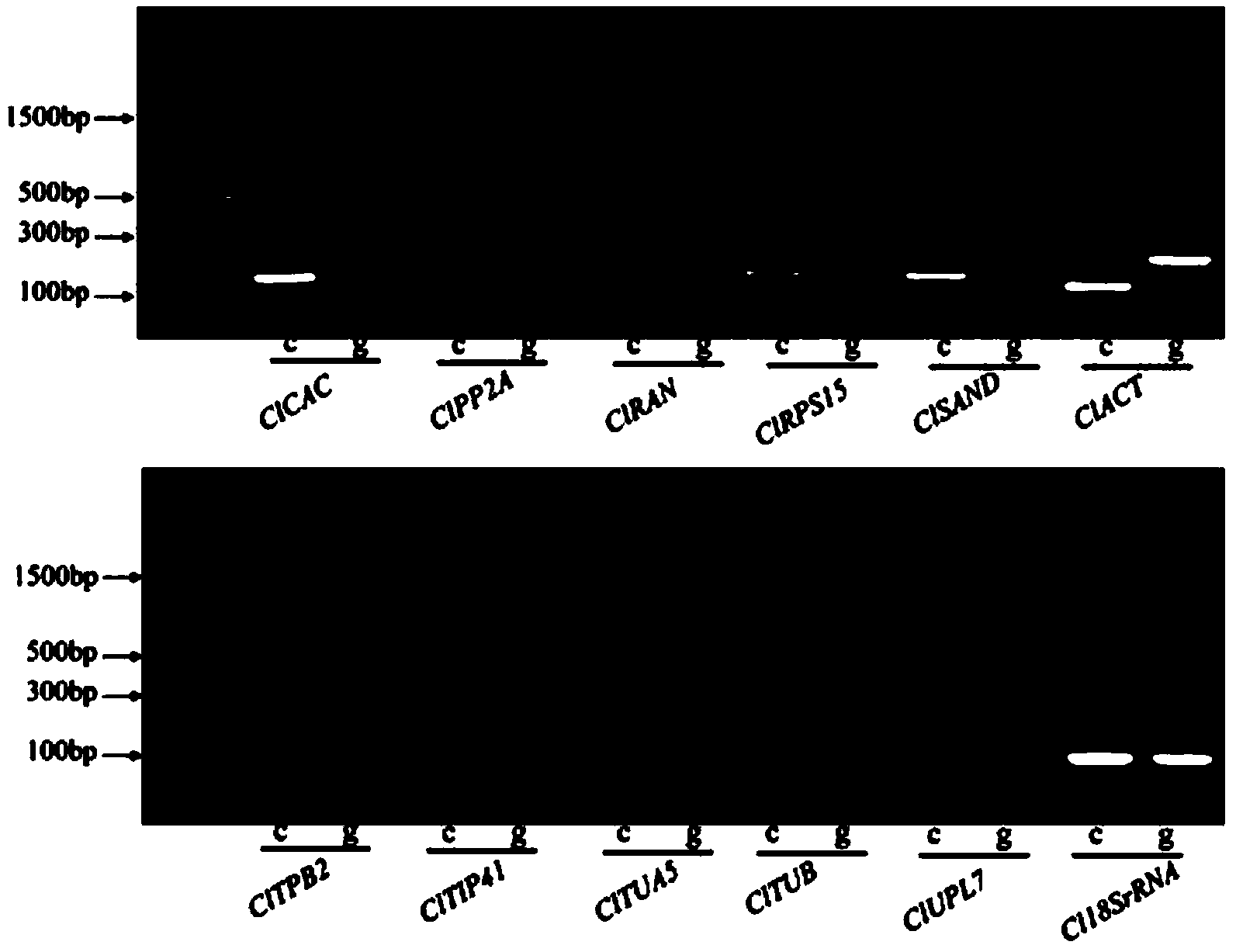
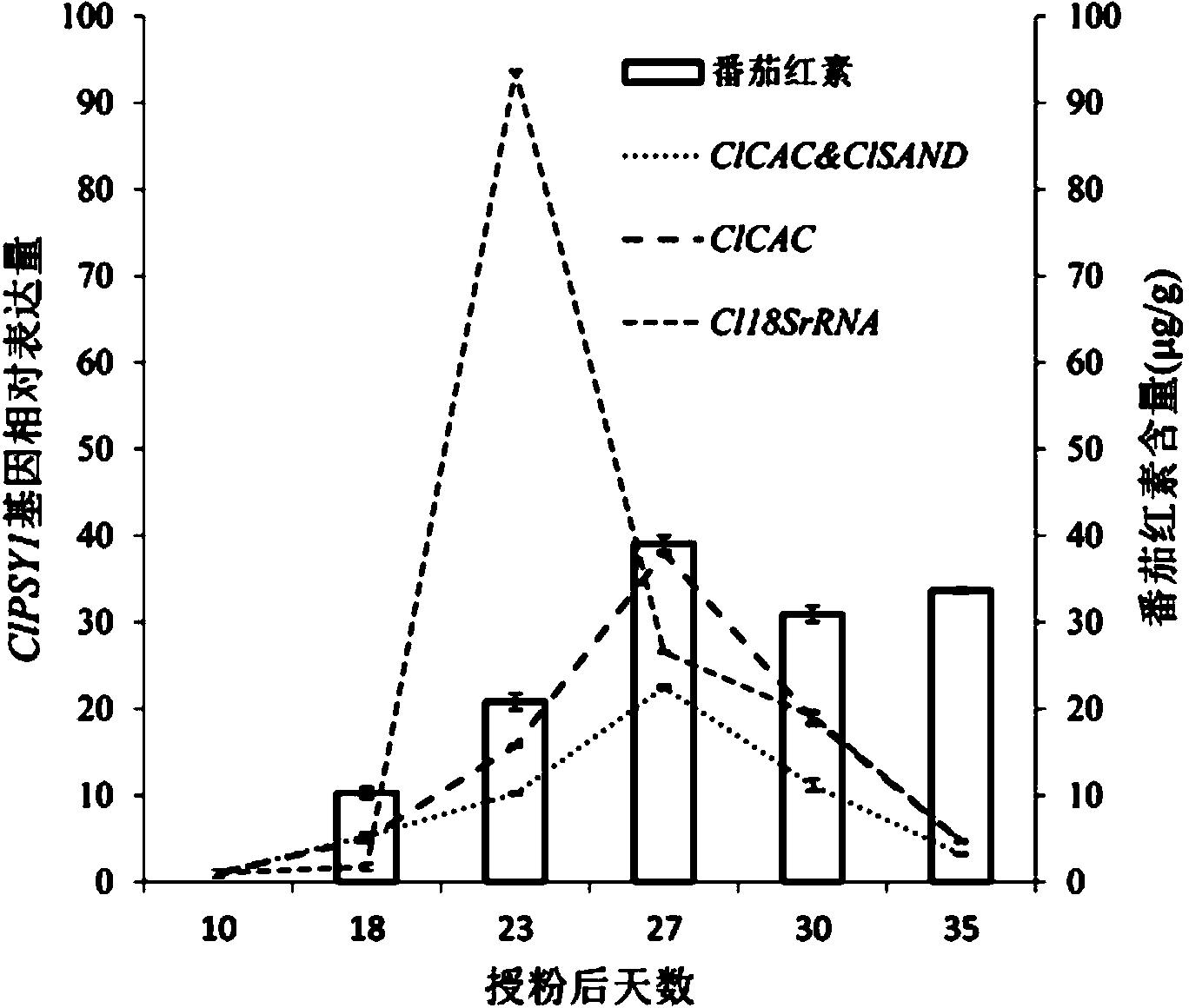
[0018] (1) Plant material and treatment: Watermelon inbred line 97103 and F1 hybrid 8424 were selected as materials, planted in plastic greenhouses, and the ovaries that were about to bloom were bagged and isolated one day before flowering. On the day of flowering, artificial pollination and CPPU treatment were used to induce watermelon fruit setting. CPPU (forchlorfenuron 0.1%; Sichuan Guoguang Agrochemical Co., Ltd.) was used at a concentration of 20 mg / L, and the paper bag was removed 8 hours in the morning, sprayed evenly on the watermelon ovary, and then bagged for isolation to prevent pollination. Each treatment was repeated 3 times and arranged randomly. Fruits were sampled at 0, 1, 3, 5, 7, 10, 12, 18, 23, 27, 30 and 35 days after pollination and CPPU treatment, and 3 fruits were randomly selected for each treatment as each sampling point biological replicates. After pollination and CPPU treatment, the ovary was taken 0, 1, 3 days after, and the center pulp was taken...
Embodiment 2
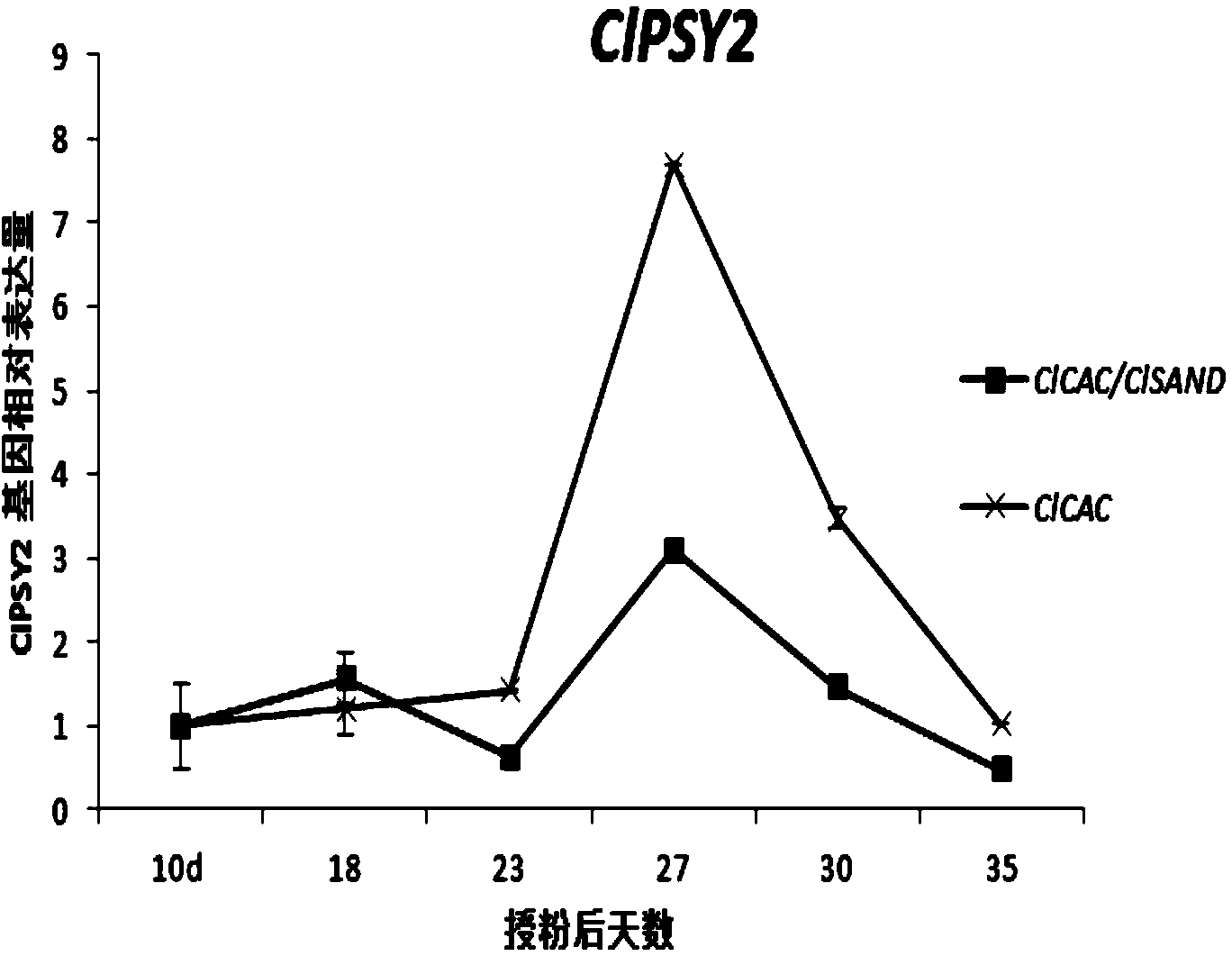
[0032](1) Plant material: the watermelon F1 generation hybrid 8424 was planted in a plastic greenhouse, and the branches were artificially pollinated and marked on the day of flowering. The fruits were sampled at 10, 18, 23, 27, 30 and 35 days after artificial pollination, and two fruits were randomly selected at each time point as a biological repeat, and three biological repeats were set up. The central pulp of the watermelon fruit was taken, quick-frozen in liquid nitrogen, and stored at -80°C.
[0033] (2) Isolation of total RNA, synthesis of first-strand cDNA and isolation of DNA were carried out according to the method described in Example 1.
[0034] (3) Target genes: Phytoene synthase gene (ClPSY2), carotene dehydrogenase gene (ClZDS) and β-carotene hydroxylase gene (ClCHYB1) in the lycopene synthesis pathway were selected as target genes , to analyze its expression during watermelon fruit ripening. The information of these genes is obtained from literature (Stefania...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com