Human papilloma virus genes, vector, strain, and expression method thereof
A human papilloma virus and gene technology, which is applied in the field of molecular biology, can solve the problems of low expression level and no expression, and achieves the effects of high expression amount, simple operation, and large-scale industrial production.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1: HPV52L1 codon optimization design
[0051] The 52L1 sequence was synthesized based on the wild-type HPV52L1 amino acid sequence (GenBank: CAA52590.1, SEQ ID NO: 1) and Pichia-preferred codons. The wild-type HPV52L1 DNA sequence was transformed, all codons were used in Pichia pastoris with higher or highest frequency of use, and the formation of secondary structure and the selection of enzyme cutting sites were considered to finally obtain the HPV52L1 gene of the present invention. Nucleotide sequence SEQ ID NO:2.
Embodiment 2
[0052] Embodiment 2: Construction of HPV52L1 recombinant expression vector
[0053] The synthesized 52L1 sequence was cloned into pPICZalphaB vector (Invitrogen) by the following method. The 52L1 DNA fragment with BstBI and KpnI at both ends was amplified by PCR, and PCR primers: forward primer: 5'CAGGTGATCTTCGAAACGATGAGTGTTTGGAGAC3'(BstBI) (SEQ ID NO: 7); reverse primer: 5'ATTGGTACCCTATTATCTTTTAACT3'( KpnI) (SEQ ID NO: 8). PCR program: cycle 30 times at 94°C for 5 minutes, 94°C for 30 seconds, 55°C for 30 seconds, 72°C for 1 minute and 50 seconds, 72°C for 10 minutes, 10°C for 10 minutes, and the operation ends. The PCR product was identified by agarose gel electrophoresis and the band at 1500bp was recovered (Qiagen gel extraction kit). The recovered fragments were digested with pPICZalphaB with BstBI and KpnI (New England Biolab), identified by agarose gel electrophoresis, and about 1500bp and 3600bp fragments were recovered respectively. After recovery, 52L1 and pPICZal...
Embodiment 3
[0054] Example 3: Construction and expression of HPV52L1 recombinant expression strain
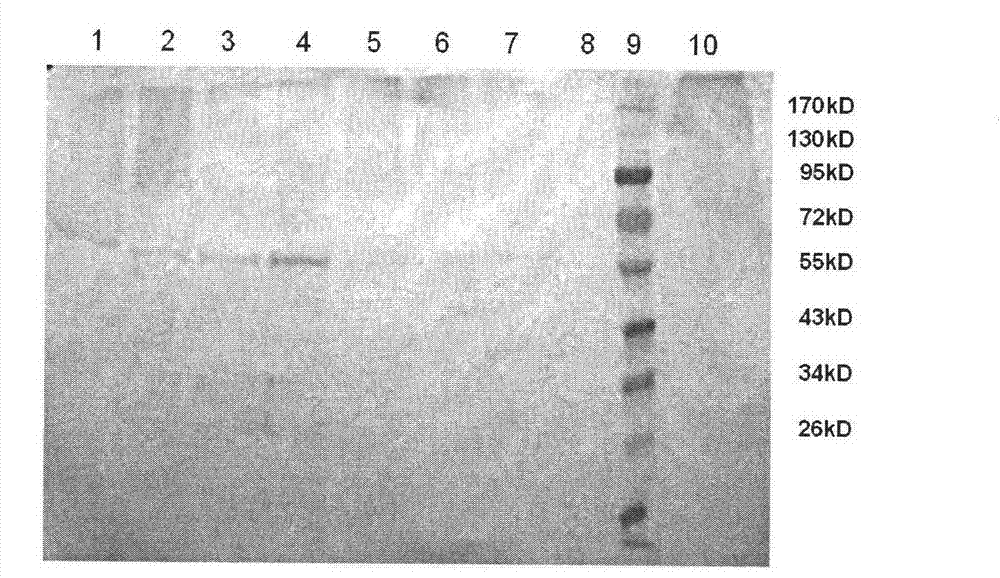
[0055] Linearize pPICZ52L1 with SacI. After the enzyme digestion reaction, remove the protein with phenol: chloroform, add 2.5 times the volume of absolute ethanol, and precipitate DNA with 1 / 10 volume of 3M NaAc (pH5.2). The resulting precipitate is washed with 75% ethanol and dried. ddH 2 O dissolved the precipitate, electroporated Pichia host bacteria (Invitrogen), spread it on a YPDS plate (containing 180 μg / mL Zeocin), and cultured at 30°C for 3 days to obtain hundreds of clones. Dozens of clones were picked and inoculated on YPD plates (containing 1500 μg / mL Zeocin), screened for high-copy plasmid strains, and cultured at 30°C for 2 days. Some clones grew faster, and several clones with the best growth conditions were picked and inoculated in 5mL YPD liquid medium. After 24 hours, the BMMY medium was replaced, and the bacteria were collected after 48 hours of induction with 0.5% met...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com