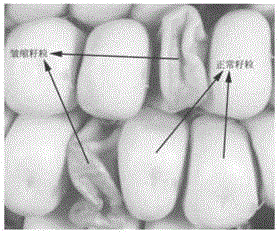
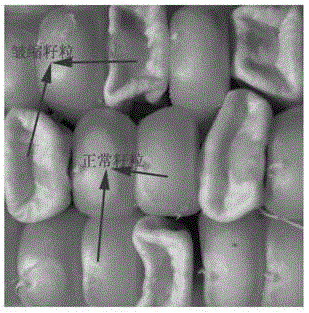
Transgenic element and application thereof, method for differentiating male sterility line and fertile maintainer line, and expanding propagation method of male sterile line of maize
A male sterility gene and genetically modified technology, applied in the agricultural field, can solve the problems of unfavorable mechanized differentiation, difference in grain shape and size, and decreased accuracy of differentiation, and achieve good application value, increase purity, and reduce volume.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] Embodiment 1 Amplification of Ms8 gene promoter sequence
[0085] The present invention takes the maize male sterile mutant Ms8 as an example to describe the embodiment in detail. By analyzing the genome sequence of Ms8, it was found that the promoter region was located 906bp upstream of the transcription start site. Taking the genome sequence of the corn inbred line B73 as a reference, the upstream and downstream primers for amplifying the promoter sequence were designed. The primer sequence Ms8-Pro-R1:5'-ggatccGCAGGCTATGCAGCTTCTCT-3' (as shown in SEQ ID NO: 7), and a Pst I restriction site was added to the 5' end of the upstream amplification primer, and the downstream amplification primer A BamH Ⅰ restriction site was added to the 5', and the length of the amplified fragment was 2343bp (Sequence ID No:2)( Figure 4 ). The PCR reaction system is (20 μl): 1 μl of B73 genomic DNA, 10.5 μl of primer Ms8-Pro-F, 0.5 μl of primer Ms8-Pro-R1, 0.5 μl of dNTP, 2 μl of 10×PC...
Embodiment 2
[0086] Example 2 Amplification of Ms8 Gene Coding Sequence
[0087] In the maizegdb database (www.maizegdb.org), the coding sequence of the Ms8 gene was found to be 1239bp (Sequence ID No: 1). Taking the sequence of the maize inbred line B73 as a reference, primers were designed to amplify the coding sequence of the Ms8 gene ( Figure 5 ), the upstream primer sequence Ms8-CDS-F1: 5'-ggatccATGCTCCAGCTGCTGCGC-3' (as shown in SEQ ID NO: 8), the downstream primer sequence Ms8-CDS-R1: 5'-gagctcTCATGTGGCGGCGTTCCA-3' (as shown in SEQ ID NO : 9), and a BamH I restriction site was added to the 5' end of the upstream primer, and a Sst I restriction site was added to the 5' end of the downstream primer. The PCR reaction system is (20 μl): 1 μl of B73 cDNA, 0.5 μl of primer Ms8-CDS-F1, 0.5 μl of primer Ms8-CDS-R1, 0.5 μl of dNTP, 2 μl of 10×PCR Buffer, 0.2 μl of Taq enzyme, ddH 2 O 15.3 μl. PCR reaction conditions: 94°C for 5min, 94°C for 35S, 58°C for 40S, 72°C for 1.5min, 35 cycles, 7...
Embodiment 3
[0088] Embodiment 3 Amplification of Bt2 gene promoter sequence
[0089] The expression of the Bt2 interference fragment in the present invention uses the promoter of the gene itself, and the expression of the Bt2 gene is mainly concentrated in the endosperm of corn seeds, so the mRNA that utilizes the promoter to initiate transcription mainly exists in the seed endosperm. The genome sequence analysis of the Bt2 gene found that the promoter region was located 1271bp upstream of the transcription initiation site. Using the genome sequence of the maize inbred line B73 as a reference, primers were designed to amplify the promoter region of the Bt2 gene. The upstream primer sequence Bt2-Pro -F1: 5'-ctgcagTTTTCTCTCTCCCGCATGTT-3' (as shown in SEQ ID NO: 10), the downstream primer sequence Bt2-Pro-R1: 5'-tctagaATGGGGAATTCGAGGAAAGT-3' (as shown in SEQ ID NO: 11), and in A Pst Ⅰ restriction site was added to the 5′ end of the upstream primer, and an Xba Ⅰ restriction site was added t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com