Escherichia coli for detecting mercury
A technology of Escherichia coli and escherichia coli, which is applied in the field of environmental biology, can solve the problems of high background of plasmid integration, low fluorescence intensity, and weak detection signal, and achieve short detection time, simple equipment and reliable results. letter effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1. Fusion of target gene
[0051] 1. Primer information and synthesis
[0052] Synthesize the fusion reporter gene and design the primer sequence according to the searched mercury-regulated gene merR and the promoter PmerT and the cfp gene sequence published in Genebank, as shown in Table 1, P 1 ,P 2 Forward and reverse primers for amplifying merR and promoter PmerT, P 3 ,P 4 Respectively amplify cfp gene forward and reverse primers, P 2 and P 3 The primer has a complementary sequence of about 20bp, and the merR+PmerT and cfp are connected in series by crossover PCR to form a fusion reporter gene. The specific information is shown in Table 1. The primers were synthesized by Shanghai Jierui Bioengineering Co., Ltd. with sterile ddH 2 O Dissolve the primers, make a storage solution with a concentration of 10 μM / L, and store at -40°C. P 5 ,P 6 and P 7 The primer connects the linker sequence to the cfp gene by PCR, P 9 ,P 10 The primers amplified the li...
Embodiment 2
[0091] Example 2. Gene knock-in
[0092] 1. Synthesis of primer information
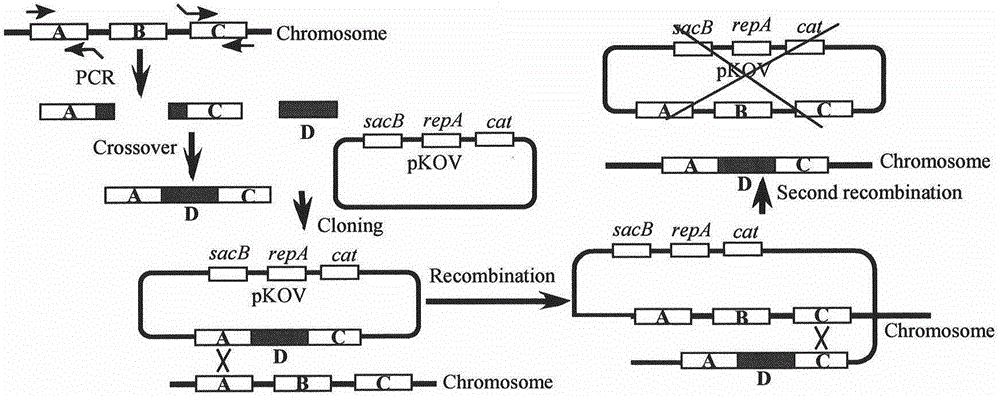
[0093] P 11-18 is the gene knock-in primer, where P 11-14 To amplify the N-terminal and C-terminal primers of zntA position respectively, P 11 and P 14 Primers contain Sal I and Not I restriction sites respectively, P 12 and P 13 Containing 20bp homology arms with the common detection element N-terminal and C-terminal respectively, P 15-16 Primers for amplifying the linearized pKOV vector at Sal I and Not I restriction sites, P 17-18 In order to amplify the primers of the mercury detection element, the mercury detection element was connected to the pKOV vector by enzyme-cut ligation, and then the mercury detection element merR::PmerT::cfp::cfp was replaced by the zntA position by gene recombination.
[0094] Table 9
[0095]
[0096] Note: The underlined gene sequence is the restriction site
[0097] 2. Amplification of two homology arms
[0098] Take 1ml of E.coli MC4100 overnight bacte...
Embodiment 3
[0153] Embodiment 3. establish the microbiological method that Escherichia coli bacterial strain E.coli WMC-010 detects mercury ion
[0154] 1 Preparation of culture medium
[0155] 1g / L tryptone, 0.5g / L yeast extract, 1g / L NaCl, first add 50ml MilliQ H 2 O, add MilliQ H after shaking until completely melted 2 O was adjusted to 80ml, 121 ° C, 30min high pressure steam sterilization, then added 10ml 400mM sterile MOPS buffer (pH 7.2), and mixed.
[0156] 2 inoculation
[0157] Take 5-10 μL of the E.coli WMC-010 bacteria solution of the recombinant strain constructed in Example 1 and add it to the 5ml fresh LB liquid medium prepared in step 1, and cultivate overnight at 37°C and 250rpm;
[0158] 3. Establishment of standard curve:
[0159] 3.1 Use a pipette gun to draw the E.coli WMC-010 bacterial solution in step 3.2, and add the calculated E.coli WMC-010 overnight bacterial seed solution to the 50ml fresh LB liquid medium prepared in step 2 to make the initial bacterial so...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com