Spider dragline silk protein optimized expression method
A technology of traction silk protein and spider silk protein, applied in the field of expression and production optimization of traction silk protein MaSp, which can solve the problem of low expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
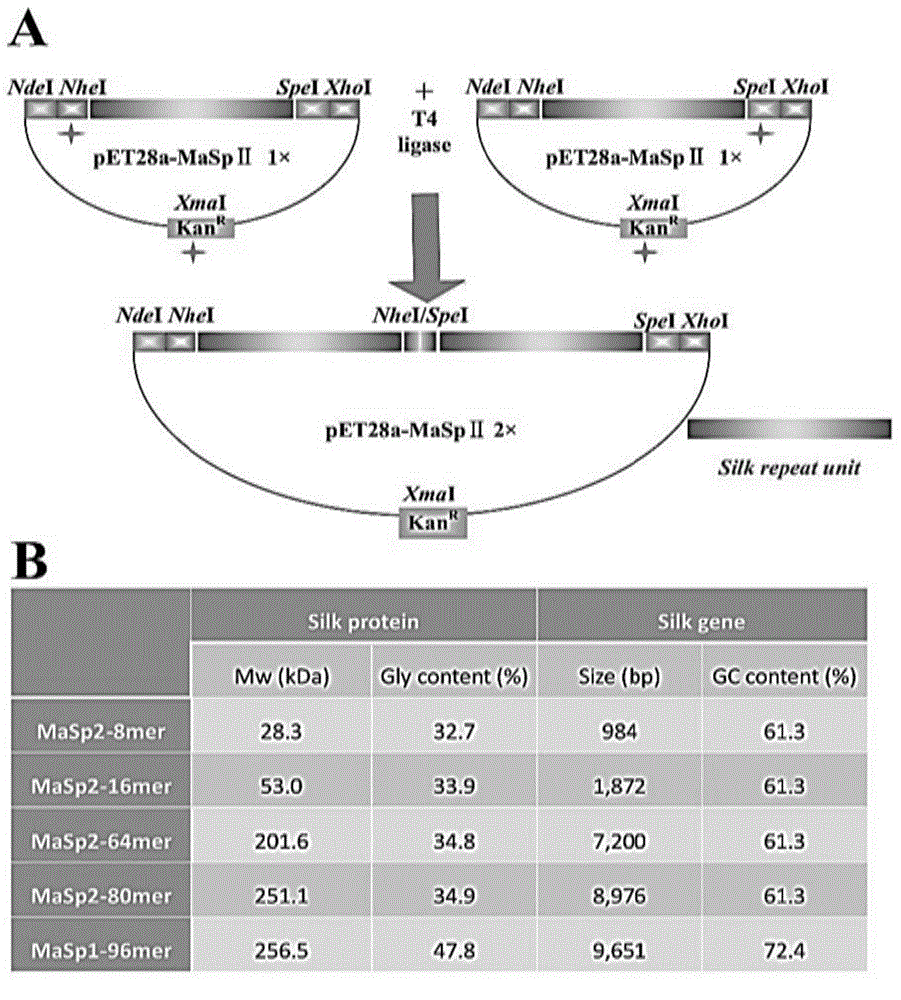
[0045] In this example, two recombinant spider silk protein expression vectors, recombinant MaSp1 and recombinant MaSp2, were respectively constructed, in which the strains, plasmids, enzymes and culture medium used were as follows: the expression plasmids were pET19b and pET-28a(+), and the expression host was large intestine Bacillus Escherichia.coli BL21 (DE3), the cloning host is Escherichia coli DH5; genetic manipulation tools include: restriction endonuclease, DNA polymerase, T4 DNA ligase; LB medium.
[0046] The component contents of the LB medium are: 10g / L tryptone, 5g / L yeast powder, and 10g / L sodium chloride.
[0047] like figure 1 As shown, the method of constructing the recombinant plasmid in this example is as follows: the strategy of "end-to-end splicing" is adopted, that is, two homologous enzymes NheI and SpeI are used for seamless splicing, and a spider with a molecular weight of 28.3kDa to 256.5kDa is synthesized. Drag silk proteins, including MaSp2-8-mer,...
Embodiment 2
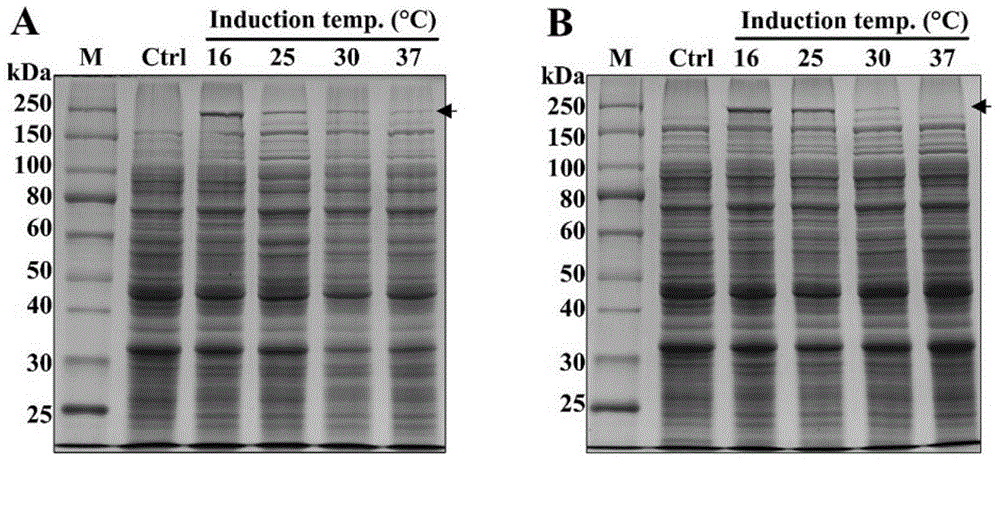
[0049] This example uses different temperatures to express and produce recombinant spider traction silk protein MaSp2-64 polymer in a shake flask system, wherein the strain is Escherichia coli BL21(DE3) / pET28a-MaSp2-64 polymer obtained in Example 1
[0050] This embodiment includes the following steps:
[0051] 1) Take out the frozen strain from -80°C, that is, Escherichia coli BL21(DE3) / pET28a-MaSp2-64 polymer, and inoculate it into 2 mL of LB medium (containing antibiotic concentration: 50 mg / mL of kanamycin), Cultivate for about 14 hours at 30°C and 220rpm.
[0052] 2) Inoculate 1 mL of seed liquid into a 250 mL shake flask containing 100 mL of LB medium or R / 2 medium at a ratio of 1:100, and culture at 30°C and 220 rpm until the initial logarithmic growth (OD 600 =0.4‐0.5);
[0053] The component content of the R / 2 medium: 2g / L diammonium hydrogen phosphate, 6.75g / L potassium dihydrogen phosphate, 0.85g / L citric acid, 5mL / L trace metal solution, 10g / L glucose, 0.7 g / L m...
Embodiment 3
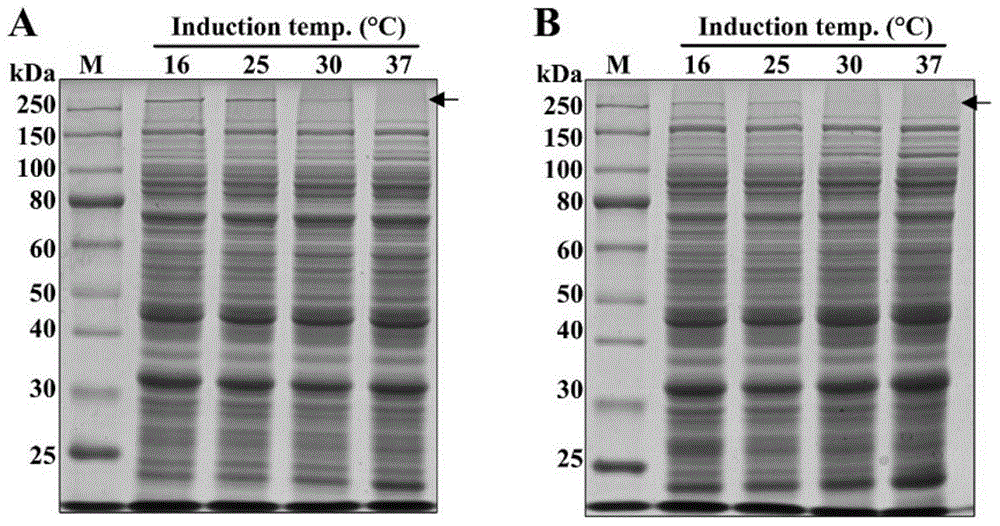
[0060] The strains used in this example are Escherichia coli BL21(DE3) / pET28a-MaSp2-80-mer and BL21(DE3) / pET19b-MaSp1-96-mer, and the seed culture and protein production fermentation operations are the same as in Example 2, wherein MaSpI96 The antibiotic concentration is 100mg / mL; Sample processing and detection operation are identical with embodiment 2, and result is as follows:
[0061] like image 3 as shown, image 3 ‐A is the protein expression gel map of MaSp2‐80mer, image 3 ‐B is the protein expression gel map of MaSp1‐96mer. After adding the IPTG initial expression factor to the recombinant engineered bacteria that were expressed and produced at 30°C, there was almost no MaSp2-80 and MaSp1-96 protein expression; while the comparison temperature culture temperature was lower than 25°C (in this embodiment, 16 ℃ and 25℃), the yields of high molecular weight recombinant spider dragline protein MaSp2-80-mer and MaSp1-96-mer can be clearly seen, and there is a significan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com