Preparation method of fusion fluorescent protein in escherichia coli
A technology of Escherichia coli and protein, applied in the direction of introducing foreign genetic material, DNA/RNA fragments, fermentation, etc. using vectors, can solve the problems of complex process and high cost, and achieve the effect of simplifying the purification process, reducing the preparation cost, and being easy to cultivate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] 1. Cloning of genes
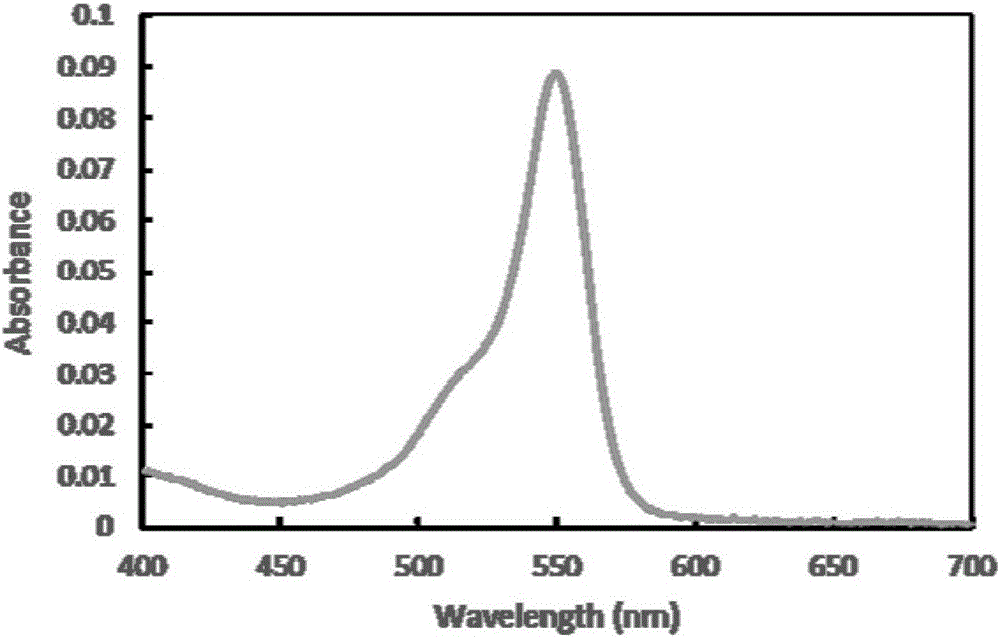
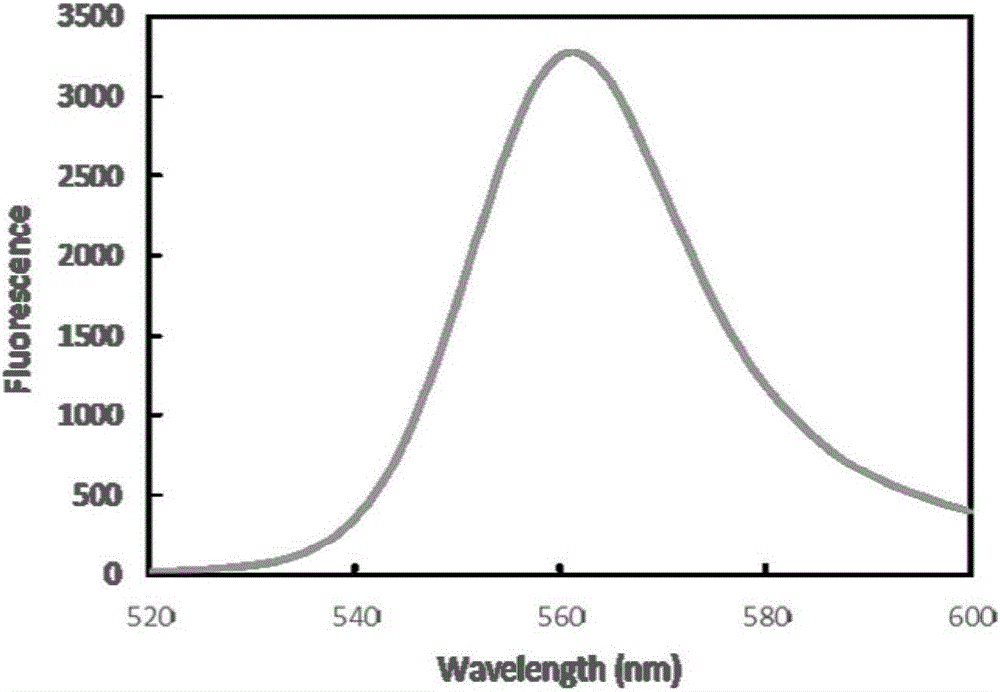
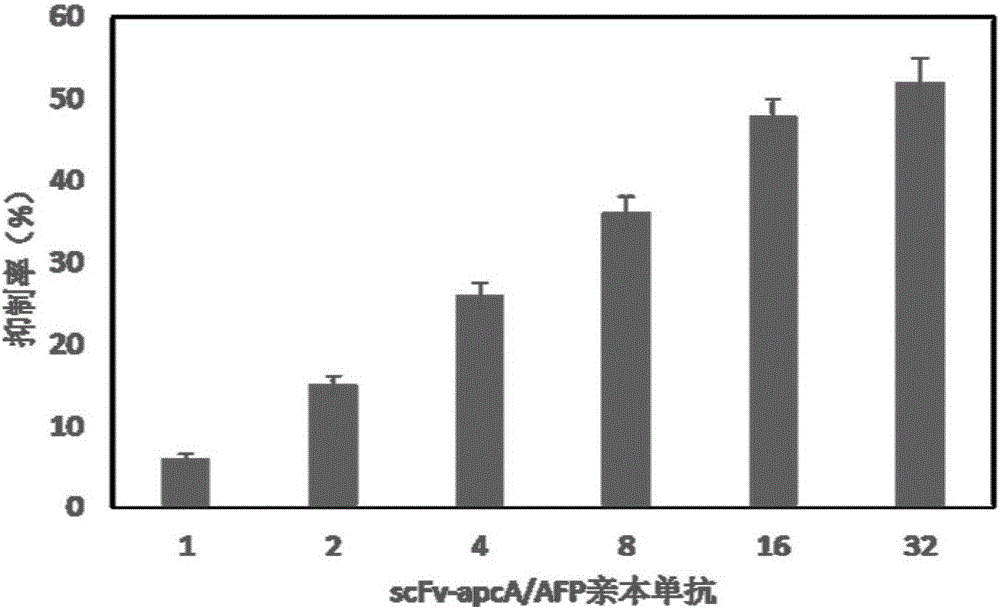
[0040] Synechococcus CC9311 apcA, Synechocytissp.PCC 6803 Ho1, Prochlorococcus phage P-SSM2 pebS, Synechococcus elongatus BP-1 cpcS and AFP single-chain antibody scFv gene sequences were obtained from the National Center for Biotechnology Information (NCBI) database (Accession No. AGQ46838). Accordingly, specific primers for amplifying apcA, Ho1, cpcS and scFv genes were designed respectively (Table 1). The apcA gene used CC9311 genomic DNA as a template, Ho1 used Synechocytissp.PCC 6803 genomic DNA as a template, cpcS used Synechococcus elongatus BP-1 genomic DNA as a template, and was amplified according to conventional PCR conditions. The pebS, scFv gene and linker sequences were artificially synthesized by Nanjing GenScript Biotechnology Co., Ltd. In order to facilitate the fusion PCR reaction, during the artificial synthesis of the linker sequence, partial sequences of the scFv gene and the apcA gene were added to its 5' end and 3' end, resp...
Embodiment 2
[0058] 1. Cloning of genes
[0059] Obtain Synechocytissp.PCC 6803 apcA, Synechocytissp.PCC 6803 Ho1, Prochlorococcus phage P-SSM2 pebS, Synechococcus elongatus BP-1 cpcS and AFP single-chain antibody scFv gene sequences from the National Center for Biotechnology Information (NCBI) database (Accession No. AGQ46838). Accordingly, specific primers for amplifying apcA, Ho1, cpcS and scFv genes were designed respectively (Table 1). Genomic DNA of Synechocytissp.PCC 6803 was used as template for apcA gene, genomic DNA of Synechocytissp.PCC 6803 was used as template for Ho1, and genomic DNA of Synechococcus elongatus BP-1 was used as template for cpcS gene, which were amplified according to conventional PCR conditions. The pebS, scFv gene and linker sequences were artificially synthesized by Nanjing GenScript Biotechnology Co., Ltd. In order to facilitate the fusion PCR reaction, during the artificial synthesis of the linker sequence, partial sequences of the scFv gene and the apc...
Embodiment 3
[0074] 1. Cloning of genes
[0075] Synechococcus elongatus BP-1 apcA, Synechocytissp.PCC 6803 Ho1, Prochlorococcus phage P-SSM2 pebS, Synechococcus elongatus BP-1 cpcS and AFP single-chain antibody scFv genes were obtained from the National Center for Biotechnology Information (NCBI) database sequence (Accession No. AGQ46838). Accordingly, specific primers for amplifying apcA, Ho1, cpcS and scFv genes were designed respectively (Table 1). Genomic DNA of Synechococcus elongatus BP-1 was used as template for apcA gene, genomic DNA of Synechocytissp.PCC 6803 was used as template for Ho1, and genomic DNA of Synechococcus elongatus BP-1 was used as template for cpcS. The pebS, scFv gene and linker sequences were artificially synthesized by Nanjing GenScript Biotechnology Co., Ltd. In order to facilitate the fusion PCR reaction, during the artificial synthesis of the linker sequence, partial sequences of the scFv gene and the apcA gene were added to its 5' end and 3' end, respect...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com