A method for separating specific metal-binding proteins by protein denaturation
A metal-binding protein and protein denaturation technology, applied in the field of protein chemistry, can solve the problems of metal-binding proteins that cannot be separated, difficult to separate metal-binding proteins, and incomplete separation, and achieve stable complexes, avoid aggregation and precipitation, and simple procedures Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] 1. Extraction of plant protein
[0035] Use 100 μmol L -1 The germinated rice seed embryos treated with copper sulfate were taken 5 days after the copper treatment, and the fresh rice seed embryos were fully ground with liquid nitrogen, then added 4 times the volume of protein extract solution, mixed well, and extracted at 4 °C for 30 min. Centrifuge at 15,000×g for 30 min at 4°C, and collect the supernatant. The denatured protein was quantified by a modified Bradford method (Ramagli, 1999) with chicken ovalbumin as the standard protein. The composition of the protein extract is: every liter of protein solution contains 50mmol / L sodium phosphate, pH 7.8, 300mmol / L sodium chloride, 0.1-1% Triton X-100 by mass percentage, 8mol / L urea, The remainder is water.
[0036] 2. Cu-NTA agarose affinity chromatography process
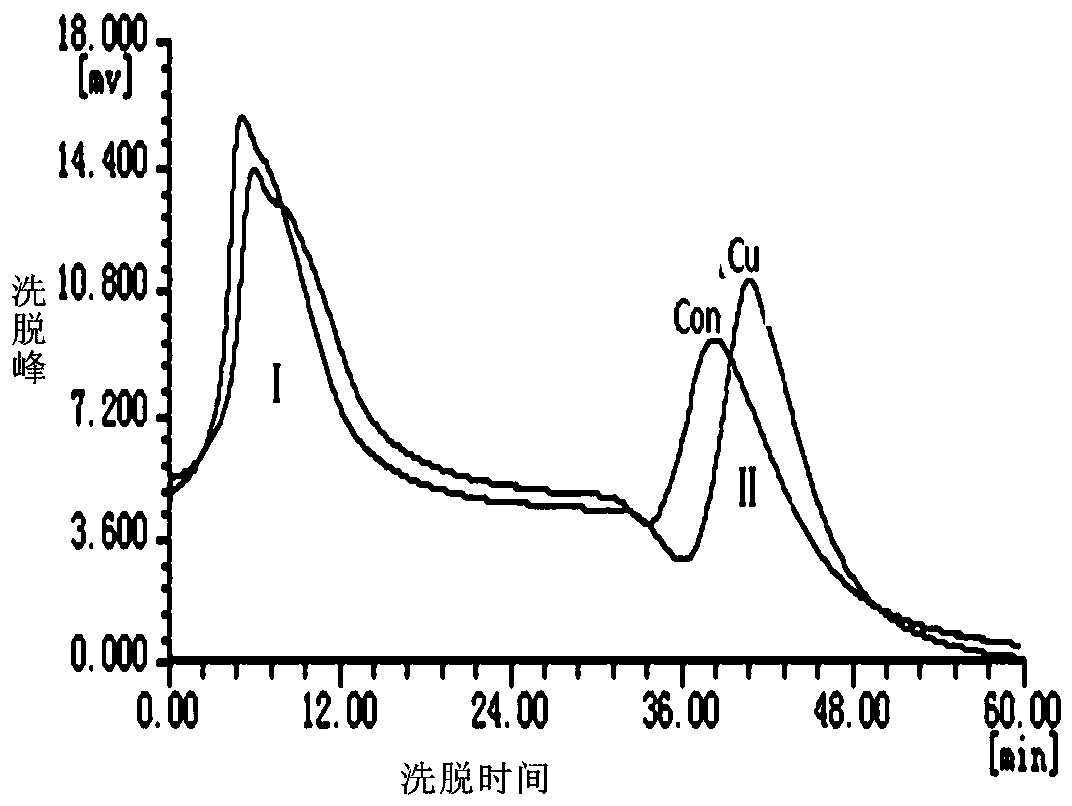
[0037] (1) Packing: Add 2 ml bed volume of Ni-NTA agarose (GE Healthcare) into the glass tube along the tube wall, and push the piston to an appropriate...
Embodiment 2
[0046] A method for separating a specific metal-binding protein by protein denaturation, comprising the following steps:
[0047] (1) Take biological tissues or cells and grind them fully with liquid nitrogen, add protein extract, mix well, and centrifuge at low temperature to obtain denatured protein solution; The solid-to-liquid ratio of the extract is: 1:4; the composition of the protein extract is: every liter of protein solution contains 50 mmol / L sodium phosphate, pH7.8, 300 mmol / L sodium chloride, and the mass percentage is 0.1 -1% Triton X-100, 8mol / L urea, the balance is water;
[0048] (2) Ni in the purchased Ni-NTA agarose (GE Healthcare) 2+ Replaced with Cu 2+ Load it into a chromatography column; inject the denatured protein solution obtained in the above step (1) into Cu-NTA agarose at a flow rate of 0.25ml / min, and incubate at 37°C for 15-40min to fully bind the denatured protein; Then, under low temperature conditions, slowly inject eluent 1 at a flow rate o...
Embodiment 3
[0057] A method for separating a specific metal-binding protein by protein denaturation, comprising the following steps:
[0058] (1) Take biological tissues or cells and grind them fully with liquid nitrogen, add protein extract, mix well, and centrifuge at low temperature to obtain denatured protein solution; The solid-to-liquid ratio of the extract is: 1:4; the composition of the protein extract is: every liter of protein solution contains 50 mmol / L sodium phosphate, pH7.8, 300 mmol / L sodium chloride, and the mass percentage is 0.2 % Triton X-100, 8mol / L urea, the balance is water;
[0059] (2) Replace the Ni in the purchased Ni-NTA agarose (GE Healthcare) with Cu and load it into the chromatographic column; inject the denatured protein solution obtained in the above step (1) into the Cu- NTA agarose, incubate at 37°C for 15-40min to fully bind the denatured protein; then slowly inject eluent 1 at a flow rate of 0.25ml / min at low temperature to refold the protein and remov...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com