Method for producing fructooligosaccharides by fermenting inulase mutants
A technology of fructooligosaccharide and inulinase, which is applied in the field of fermentation engineering, can solve problems such as complex process, high production cost, and incomplete reaction, and achieve the effects of reducing operation difficulty, saving economic cost, and reducing production cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Embodiment 1: Cloning and codon optimization of inuI gene
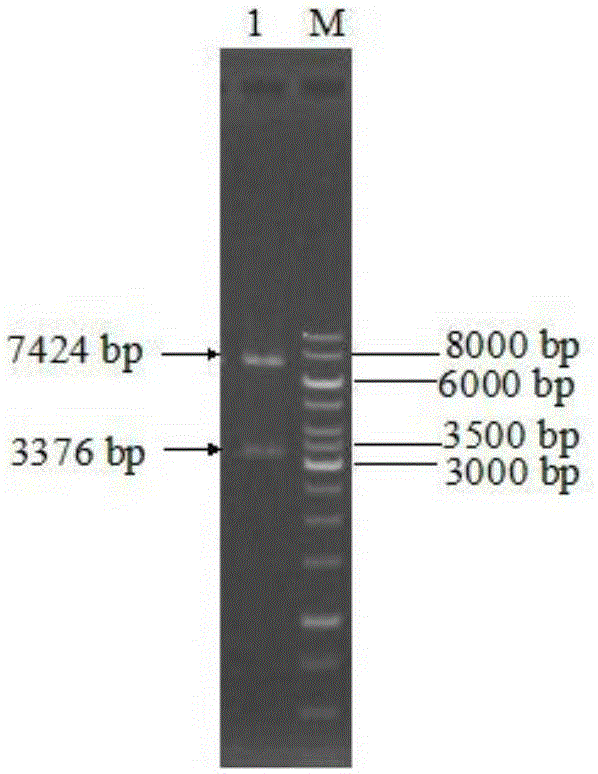
[0031] Construction of recombinant plasmid: Total RNA of Aspergillus niger NRRL3135 was extracted with TRNzol total RNA extraction reagent. Using total RNA as a template, referring to the instructions of the RT-PCR kit, using oligo(dT) as a primer to synthesize the first-strand cDNA by reverse transcription, and then respectively using the first-strand cDNA as a template, primers F1 (SEQ ID NO: 6) and R1 ( SEQ ID NO: 7) PCR amplifies the Aspergillus niger inulinase gene inuI. The PCR product and plasmid pPIC9K were digested with EcoRI and NotI, purified, ligated, and transformed into EscherichiacoliJM109 competent cells. Positive transformants were screened, and their plasmids were extracted for enzyme digestion verification. The correct recombinant plasmid verified by enzyme digestion was then sequenced (sequence SEQ ID NO: 1), and the correct recombinant plasmid constructed was named pPIC9K-inuI.
[0032] ...
Embodiment 2
[0033] Example 2: Construction of Aspergillus niger inulinase mutation library using error-prone PCR method
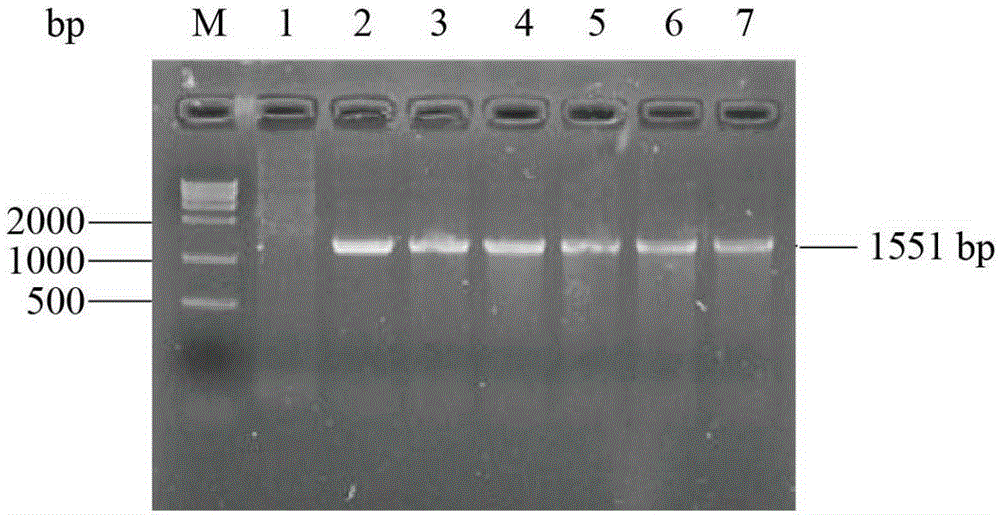
[0034] The nucleotide mutation ( figure 1 ). The reaction conditions for error-prone PCR are as follows:
[0035]
[0036] Wherein, primers F2 (SEQIDNO:8) and R2 (SEQIDNO:9) sequences (5'-3') are respectively:
[0037] Upstream primer F2: CCG GAATTC CAGTCTAATGATTACCGTCCTTCATAC
[0038] Downstream primer R2: ATAAGAAT GCGGCCGC TCATTCAAGTGAAACACTCCGC
[0039]PCR amplification conditions: 94°C for 3 min; 30 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1.5 min; 72°C for 10 min.
[0040] After the error-prone PCR amplification product is purified by DNA purification and recovery kit, it is digested with restriction endonucleases EcoRI and NotI, connected with the corresponding digested plasmid pPIC9K, and transformed into E.coliJM109 competent cells , spread on LB solid medium plate containing 100 μg / mL ampicillin. After culturing at 37°C and 200 rpm f...
Embodiment 3
[0043] Example 3: Screening of High Enzyme Activity Inulinase Mutants
[0044] Use a sterilized toothpick to remove the His grown on the MD plate + The transformant is copied to the same position of the YPD and the BMMYI plate, and the control bacteria GS115 / pPIC9K-inuI' (without error-prone PCR, the preparation process is the same as the product of Example 2) is inoculated on the BMMYI plate. Incubate at 30°C for 2 days, and save the grown YPD plate.
[0045] Plate primary screening: Induce the mutant strains on the BMMYI plate for 2-3 days. The strain with the transparent circle around the bacterium colony on the plate larger than the control bacterium GS115 / pPIC9K-inuI' is the mutant strain of the primary screening purpose.
[0046] 96-well plate re-screening: Add 300 μL of BMGY medium to a 1.8 mL / well (flat-bottomed) 96-well plate, and sterilize at 121° C. for 20 minutes. Into it, insert the primary screening target strain preserved on the YPD plate (simultaneously inse...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com