Bacillus licheniformis engineering bacterium capable of efficiently secreting nattokinase
A technology of Bacillus licheniformis and nattokinase, which is applied in the fields of enzyme engineering and microorganisms, and can solve the problems such as the lack of universality of signal peptides
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
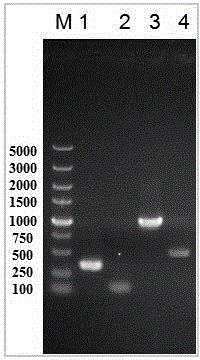
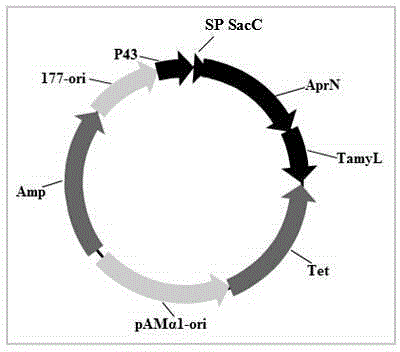
[0031] Example 1 Construction of nattokinase-producing engineered strain Bl10 (pP43SacCNK) from Bacillus licheniformis
[0032] According to the genome sequence of Bacillus subtilis 168 published by NCBI, the sequence of promoter P43 and the signal peptide sequence of fructanase SacC were amplified by PCR. The primers are:
[0033] P43-F: CGGAATTCTGATAGGTGGTATGTTTTCG
[0034] P43-R: CTTGAATCAGTCTCTTTTTTCATTTCATGTGTACATTCCTCTC
[0035] Sp-SacC-F:GAGAGGAATGTACACATGAAATGAAAAAGAGACTGATTCAAG
[0036] Sp-SacC-R: CTGTACTGCTTTTTCCGGCTGCATCTGCCGAAAATGCCATAG
[0037] According to the genome sequence of Bacillus subtilis natto published on NCBI, the nattokinase gene was amplified by PCR wxya sequence. Primers are as follows:
[0038] AprN-F: CTATGGCATTTTCGGCAGATGCAGCCGGAAAAAAGCAGTACAG
[0039] AprN-R: ATCCGTCCTCTCTGCTCTTTTGTGCAGCTGCTTGTACGTTGAT
[0040] According to the genome sequence of Bacillus licheniformis WX-02 published on NCBI, the amylase gene was amplified by PCR am...
example 2
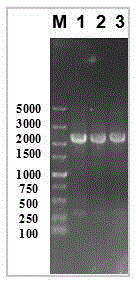
[0043] Example 2 Fermentation experiment and enzyme activity determination of Bacillus licheniformis Bl10 (pP43SacCNK)
[0044] Seed solution: 10g / L peptone, 5g / L yeast extract powder, 10g / L sodium chloride pH7.2-7.4 250mL triangular bottle liquid volume is 50mL
[0045] Seed cultivation time: 10h
[0046] Fermentation medium: 20g / L glucose, 10g / L peptone, 15g / L yeast extract powder, 10g / L sodium chloride, 6g / L ammonium sulfate; 10g / L soybean peptone, 5g / L corn steep liquor pH7.0-7.2 250mL triangular bottle The volume is 30mL
[0047] Inoculum size: 3%
[0048] Training time: 48h
[0049] Fermentation broth pretreatment: Take 2mL of fermentation broth in a 2mLEP tube, centrifuge at 10000rpm for 5min, transfer the supernatant to another 2mLEP tube, and store at 4°C for later use.
[0050] Determination of enzyme activity by fibrinogen plate method: At 50°C, 10 mL of fibrinogen solution (2.0 mg / mL) and 10 mL of agarose solution (1.0%, w / v) were quickly mixed,...
example 3
[0051] Example 3 Determination of nattokinase secretion at the end point of fermentation of Bacillus licheniformis Bl10 (pP43SacCNK)
[0052] Preparation of BSA standard curve: BSA solutions of different concentrations (0.2mg / mL, 0.4mg / mL, 0.6mg / mL, 0.8mg / mL, 1mg / mL) were mixed with the same amount of 2*SDS-PAGEbuffer, and then 10ul samples were taken. After electrophoresis, Obtain the standard curve of BSA by detecting the depth and area of its different concentrations of BSA bands, which is: y=1.3522x+0.7429 (R 2 =0.9955, y: depth area product of protein band x: total amount of protein)
[0053] Fermentation broth sample pretreatment: According to the method, pretreat the supernatant of the fermentation broth, mix it with 2*SDS-PAGEbuffer, take 10ul spotting, and compare the color depth and area of the protein band with the standard curve of BSA to obtain The protein content of nattokinase in the fermentation broth was 163.52mg / L.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com