Tanshinone biosynthesis inhibiting factor gene SmJAZ3 and encoding protein and application thereof
A technology for biosynthesis and inhibitory factors, applied in the field of genetic engineering, which can solve the problems that have not yet been discovered and that there are few researches on the signal transduction of Salvia miltiorrhiza
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Cloning of Example 1 Danshen SmJAZ3 Gene
[0022] 1. Tissue separation (isolation)
[0023] The tender tissues of Salvia miltiorrhiza plants were taken and stored in -80°C refrigerator
[0024] 2. RNA isolation (RNAisolation)
[0025] Take part of the tissue and grind it with a mortar, add the lysate to homogenize it, transfer it to a 1.5mLEP tube, and extract the total RNA. Ordinary agarose gel electrophoresis was used to identify the quality of total RNA, and then the RNA content was determined on NanoDrop.
[0026] 3. Cloning of Full-length cDNA
[0027] The hairy roots of Salvia miltiorrhiza in shake flasks cultured for two months were induced by methyljasmonic acid, RNA was extracted, reverse transcribed into cDNA, and transcriptome analysis was performed by high-throughput sequencing.
[0028] 3.1. Transcriptome sequencing
[0029] Through transcriptome sequencing of the hairy root material of Salvia miltiorrhiza, a highly homologous sequence to Arabidopsis J...
Embodiment 2
[0035] Example 2 (Sequence information and homology analysis of Danshen SmJAZ3 gene)
[0036] The length of the full-length cDNA of the SmJAZ3 gene of the present invention is 1011bp, and the detailed sequence is shown in SEQ ID NO.1. The amino acid sequence of the Danshen SmJAZ3 gene was deduced according to the full-length cDNA, with a total of 336 amino acid residues. See SEQ ID NO.2 for the detailed sequence.
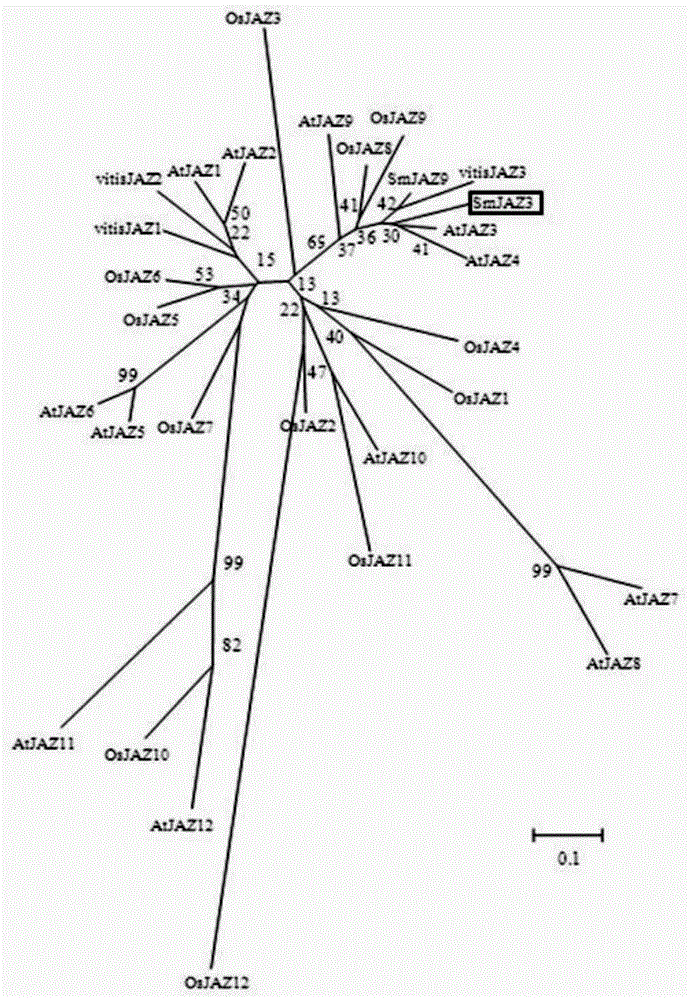
[0037] The homologous comparison of the amino acid sequences of Salvia miltiorrhiza SmJAZ3 and tobacco (Nicotianatabacum) jasmonateZIM-domainprotein7b is shown in Table 1. The gene was compared with the Arabidopsis JAZ gene by software such as ClustalW and Boxshade. This gene has the conserved ZIM structure of AtJAZs and the Jas domain (SLX2FX2KRX2RX5PY). The result is as Figure 10 , Figure 11 shown. The phylogenetic tree was established by Mega6.0, and the local Blast found that the obtained JAZ gene had the highest homology with Arabidopsis JAZ3, and named i...
Embodiment 3
[0056] Example 3 Construction of overexpression and antisense suppression vectors, genetic transformation of Salvia miltiorrhiza hairy roots
[0057] 1. Construction of SmJAZ3 overexpression and antisense suppression vector
[0058] SmJAZ3 forward and reverse full-length clones into pCAMBIA2300 + On, the overexpression vector pCAMBIA2300 was obtained + -SmJAZ3 and antisense suppression vector pCAMBIA2300 + -Anti-SmJAZ3.
[0059] 2. Agrobacterium rhizogenes C58C1 mediates pCAMBIA2300 + , pCAMBIA2300 + -SmJAZ3 and pCAMBIA2300 + -Anti-SmJAZ3 plasmid genetic transformation of Salvia miltiorrhiza to obtain hairy roots of Salvia miltiorrhiza.
[0060] (1) Pre-cultivation of explants
[0061] Cut the leaves and petioles of sterile salvia miltiorrhiza seedlings, inoculate them on the preculture medium (1 / 2MS), and culture them in the dark at 25°C for 2 days.
[0062] (2) Co-cultivation of Agrobacterium and explants
[0063] Put the above-mentioned explants of pre-cultivated l...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com