A human dipeptidyl peptidase III modified by site-directed mutagenesis
A technology of dipeptidyl peptidase and site-directed mutagenesis, applied in the direction of peptidase, enzyme, hydrolase, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: Synthesis of wtDPP and humDPP
[0025] The present invention takes the dipeptidyl peptidase gene (NCBI database AJ271216.1) sequence of Homo sapiens as a reference, and adds 5'-GTC at the 5' end GAATTC -3' join 3'- at the 3' end CCTAGG GAC-5', ( GAATTC ) is the restriction enzyme cutting site EcoRI, ( GGATCC ) is the restriction enzyme cutting site BamHI. The wtDPP gene was synthesized by artificial total synthesis.
[0026] The present invention takes the dipeptidyl peptidase gene (AJ271216.1) sequence of Homo sapiens as a reference, the 560th amino acid is substituted with alanine (Alanine, ALa, A), and the 562nd amino acid is substituted with lysine ( Lysine, Lys, K) substitution, the 564th amino acid is replaced by histidine (Histidine, His, H), and 5'-GTC is added at the 5' end GAATTC -3' join 3'- at the 3' end CCTAGG GAC-5', ( GAATTC ) is the restriction enzyme cutting site EcoRI, ( GGATCC ) is the restriction enzyme cutting site BamHI. ...
Embodiment 2
[0029] Example 2: Construction of recombinant plasmids wtDPP and humDPP expression plasmids
[0030] Gene cloning was carried out according to conventional methods (Sambrook, et al. 2001, Molecular Cloning A Laboratory Manual. Cold Spring Harbor Laboratory Press. USA), and the wtDPP and humDPP obtained in Example 1 were respectively cloned into pHIL-S1 to construct the expression plasmid pHIL-S1 -wthDPP and the expression plasmid pHIL-S1-humDPP, the cloned target gene was identified by enzyme digestion and sequencing.
[0031] specific methods:
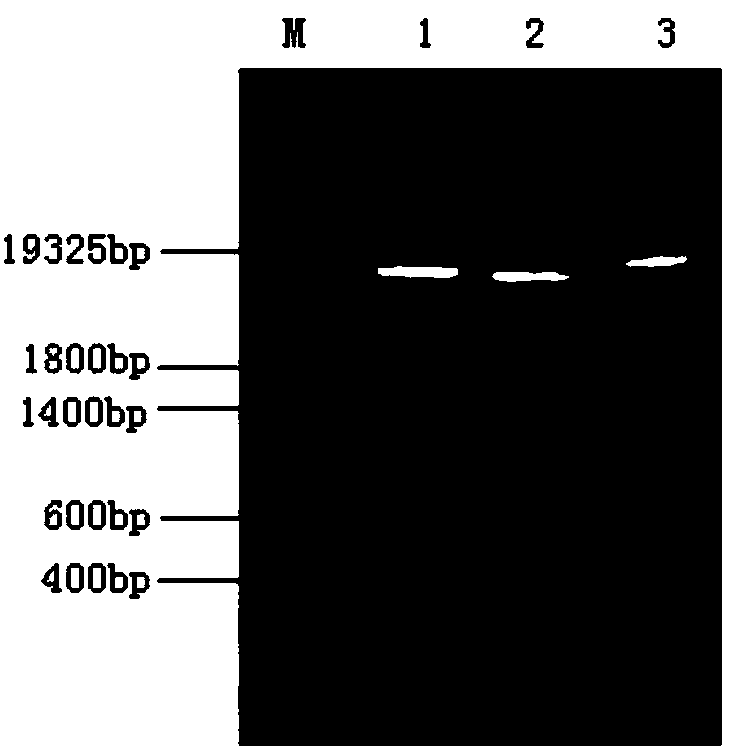
[0032] The construction process of the recombinant plasmid pHIL-S1 containing humDPP is as follows: EcoRI+BamHI double-digestion of the plasmid pHIL-S1 and the target fragment humDPP, the digested products were subjected to 0.8% agarose gel electrophoresis, and the gel was cut and recovered. Ligation of plasmid pHIL-S1 and humDPP was performed using T4 DNA ligase. CaCl 2 Prepare E.coli DH5α competent cells, transform DH5α competent...
Embodiment 3
[0034] Example 3: Expression of recombinant humDPP and recombinant wthDPP
[0035] Expression of recombinant humDPP: Recombinant plasmid pHIL-S1-humDPP and plasmid pHIL-S1 were digested with SacI, the digested products were subjected to 0.8% agarose gel electrophoresis, and the linear recombinant plasmids pHIL-S1-humDPP and Plasmid pHIL-S1. According to the protoplast method in the Pichia Expression Kit (Invitrogen Inc., U.S.) manual, Pichia hominis GS115 was transformed, and Mut + Turn. Using methanol as the sole carbon source to induce expression of recombinant bacteria (operated according to the PichiaExpression Kit manual), the SDS-PAGE electrophoresis analysis of the culture solution showed that after the transformants were induced to express, the supernatant of the culture solution showed obvious target protein bands, while The control bacteria transformed with an empty plasmid without the target gene were induced under the same conditions for 96 hours, and no target p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com