Plasmid carrier construction method applied to Shewanella
A technology of Shewanella and plasmid vectors, which is applied in the field of construction of plasmid vectors for Shewanella, and can solve problems such as lack of and no major breakthroughs in Shewanella
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Embodiment 1: the cultivation of thalline
[0030] 1) Solid culture
[0031] The composition of LB medium: tryptone 10g / L, yeast extract 5g / L, sodium chloride 10g / L and agar 15g / L, streak inoculated on solid medium. Shewanella xiamenensis BC01 and Shewanellaoneidensis MR-1 were cultured at 30°C for 16h, and Escherichia coli DH5α were cultured at 37°C for 16h. If the bacterium carries the kanamycin resistance gene, the culture medium needs to add 1% kanamycin.
[0032] 2) Liquid culture
[0033] First, pick a single colony from the LB agar plate with an inoculation loop and inoculate it into a 250mL Erlenmeyer flask containing 50mL of LB culture solution. Shewanella xiamenensis BC01 and Shewanellaoneidensis MR-1 were cultured at 30°C for 16h, and Escherichia coli DH5α were cultured at 37°C for 16h. If the bacteria have a kanamycin resistance gene, the culture medium needs to be added with kanamycin, and the final concentration is 500mg / ml.
Embodiment 2
[0034] Embodiment 2: the extraction of plasmid
[0035] 1) Initial extraction of plasmids
[0036] Plasmid DNA was extracted from cultured Shewanella xiamenensis BC01 using OmegaPlasmidMiniKit kit. See the kit instructions for details.
[0037] 2) Agarose gel electrophoresis
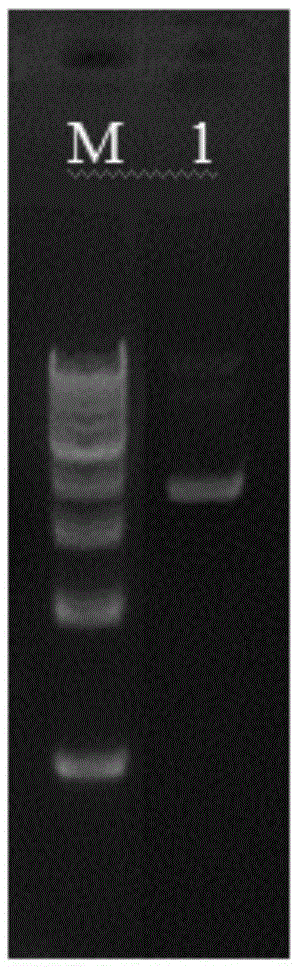
[0038] Add 25ml of 1×TAEbuffer and 0.25g of agarose into the beaker. Heat it in a microwave oven to dissolve it completely, wait for it to cool to about 60°C, pour it into a cleaned glue-making tank, and insert a 50μL glue-making comb. After cooling and solidification, carefully pull out the comb. Put the gel together with the gel-making tank in the electrophoresis tank equipped with 1×TAEbuffer, with the side with the hole facing the negative electrode, and the liquid level of 1×TAEbuffer should completely submerge the gel. Take an appropriate amount of DNA sample, that is, mix the extracted pSXM33 and 6xloadingbuffer at a ratio of 5:1, add it to the sample well, and finally add 5-10 μL DNAmarker. ...
Embodiment 3
[0041] Example 3: Restriction Enzyme Digestion and Verification of Plasmids
[0042] In this experiment, fast cutting enzyme was used for enzyme digestion, and the enzyme digestion system was reacted in a water bath at 37° C. for 30 minutes, and then verified by agarose gel electrophoresis. The electrophoresis steps were the same as those described in Example 2.
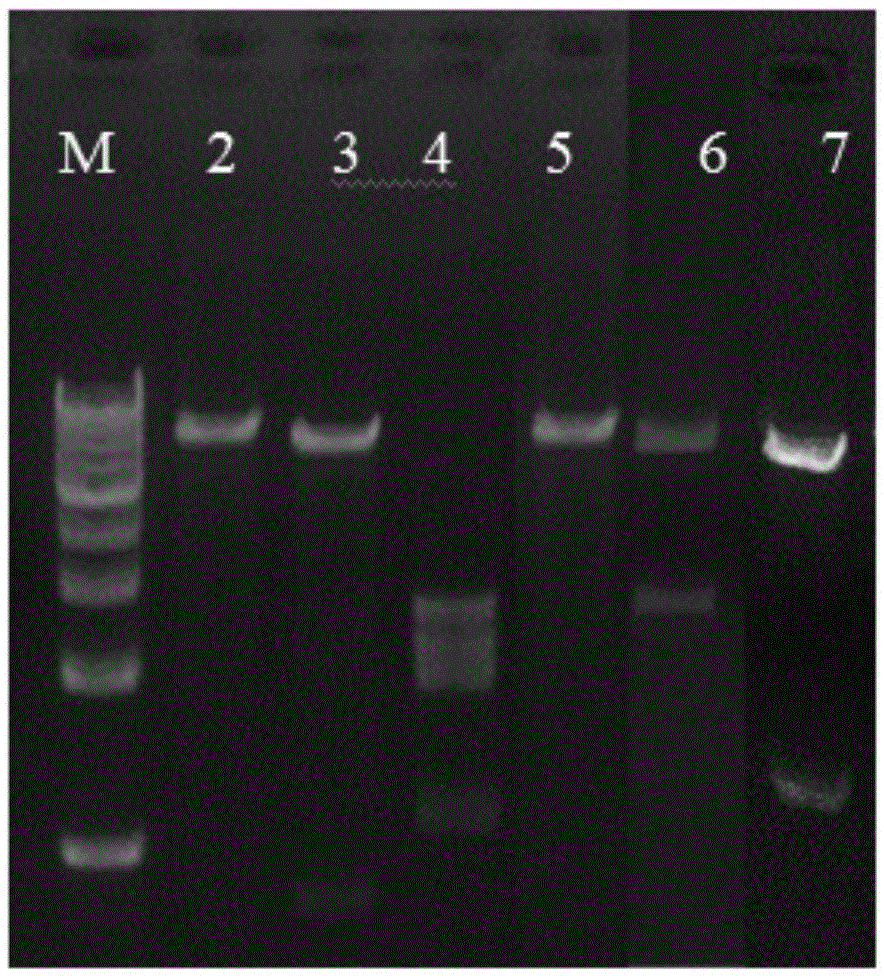
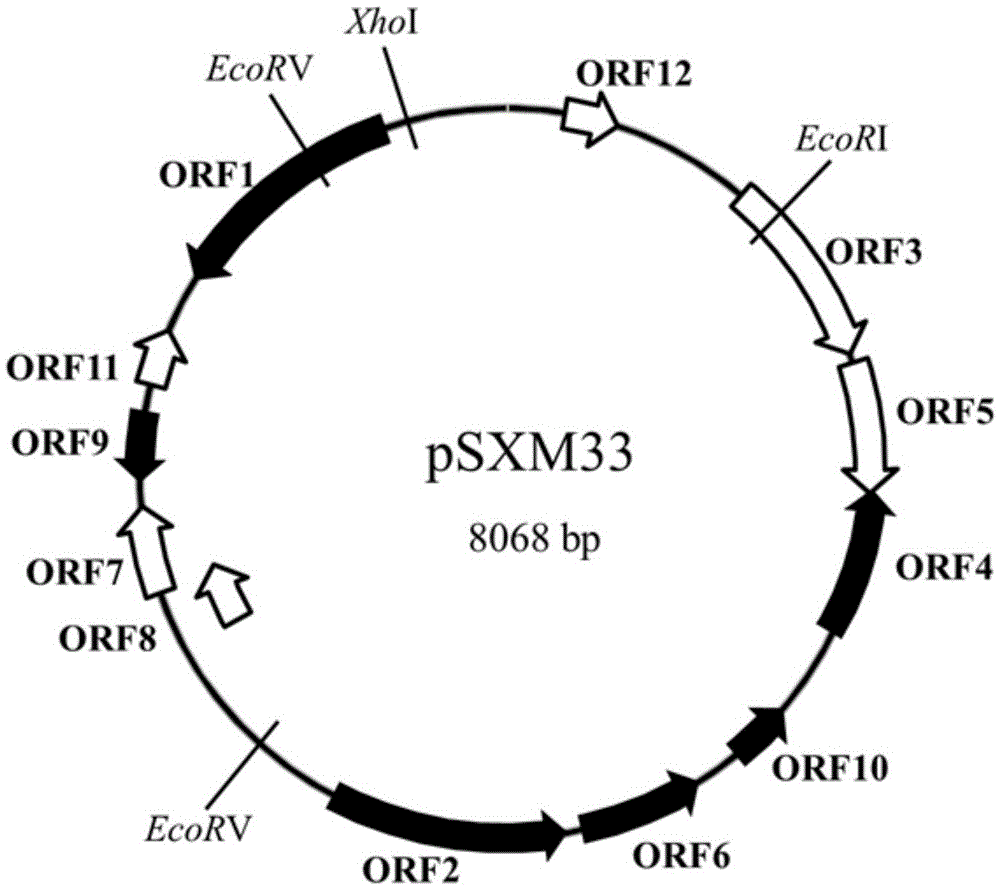
[0043] The verification scheme is Lane2:XhoI, Lane3:HindIII, Lane4:PstI, Lane5:EcoRI, Lane6:EcoRV, Lane7:XhoI+EcoRI. After trying the above enzyme digestion method, the result is as follows figure 2 As shown, there are EcoRV, EcoRI and XhoI restriction enzyme sites in the pSXM33 plasmid sequence, and each site is unique on the plasmid.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com