Method and apparatus for determining fetus target area haplotype
A target region and haplotype technology, applied in the field of bioinformatics, can solve the problem of low fetal DNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
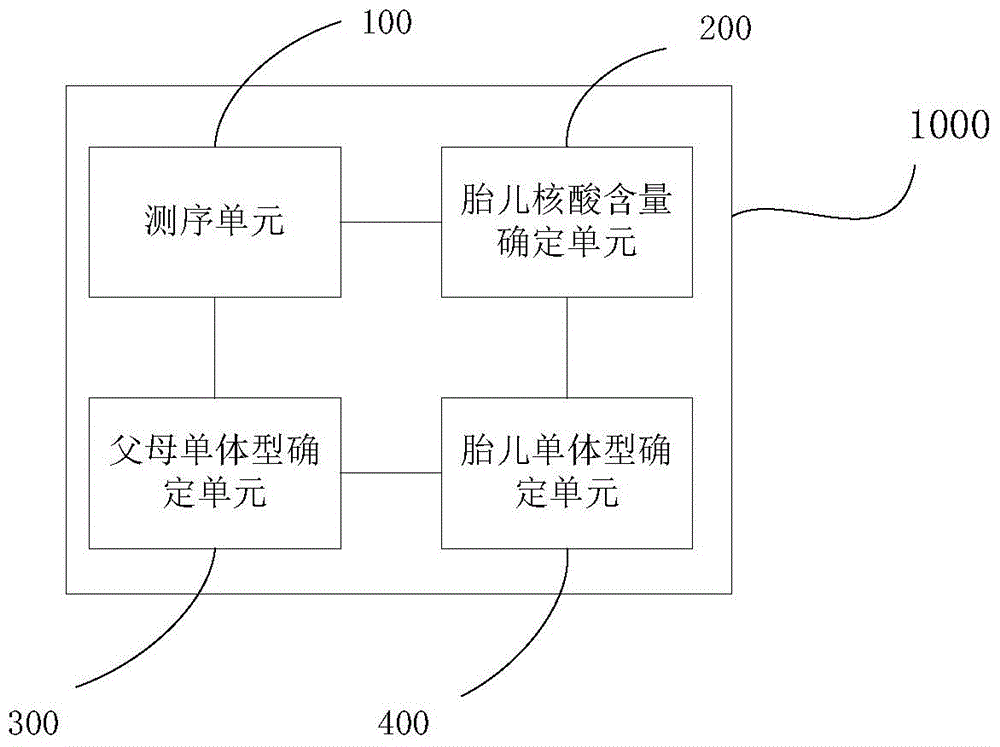
[0012] According to one embodiment of the present invention, a method for determining the haplotype of a fetal target region is provided, comprising the following steps:
[0013] Step 1: Obtain the first, second, third and fourth sequencing data.
[0014] The free nucleic acid in the body fluid of the pregnant woman is obtained, the target region is captured, and sequence determination is performed on the captured target region to obtain first sequencing data. The body fluid sample of a pregnant woman is a sample containing fetal nucleic acid. For example, the peripheral blood plasma of a pregnant woman contains fetal nucleic acid, and the extracted peripheral blood free nucleic acid is a mixture of nucleic acid of the pregnant woman and the fetus, and the mixture is highly fragmented. According to the existing sequencing platform, by constructing the sequencing library of the free nucleic acid extracted from the peripheral blood samples of pregnant women, using probes or ch...
Embodiment
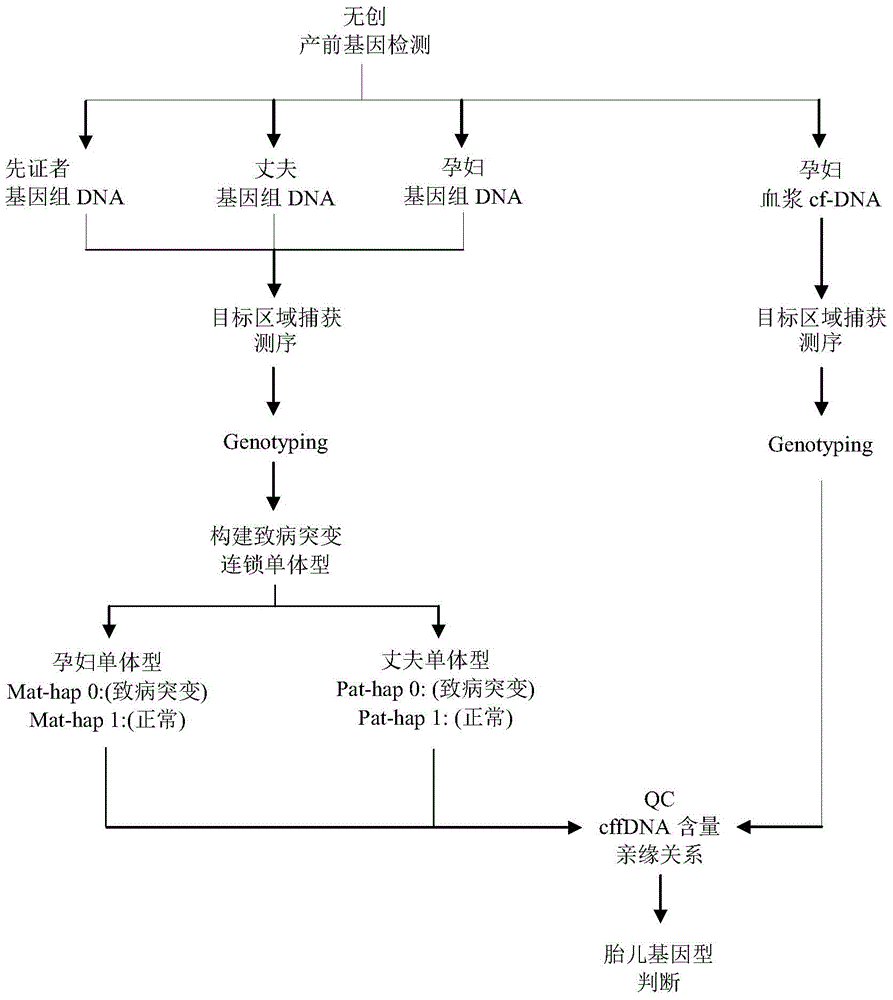
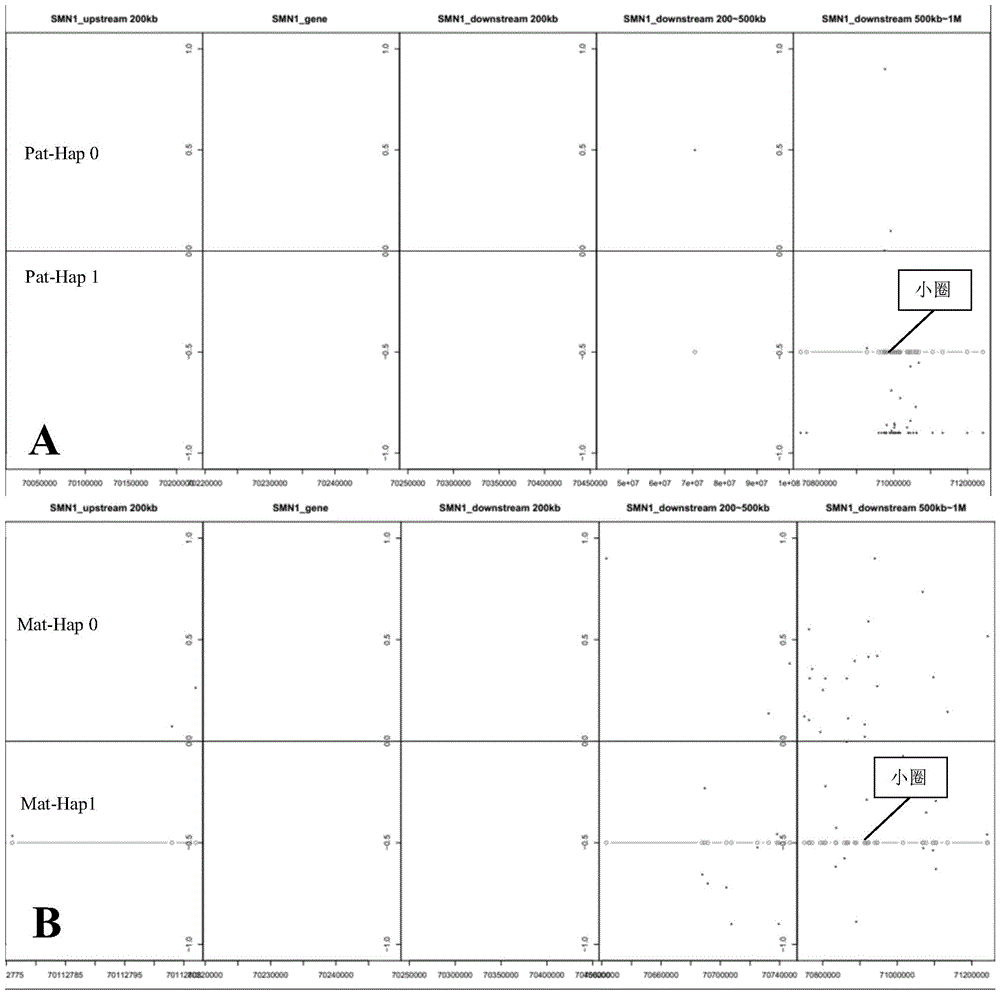
[0065] Non-invasive prenatal genetic testing was performed on a pregnant woman with a high risk of giving birth to a second child with SMN1 disease (Tianjin Maternal and Child Health Hospital). Both the pregnant woman and her husband were heterozygous carriers for deletion mutation of exon 7 of SMN1 gene, and had given birth to a patient with homozygous mutation of SMN1. Now that she is pregnant for the second time, the peripheral blood of the pregnant woman is drawn and the plasma is separated in time, and then the DNA of the plasma DNA and the genomic DNA of the pregnant woman, pregnant woman's husband, and proband are captured and sequenced to analyze the genetic status of the fetus.
[0066] The DNA of the sample is extracted by the salting out method, and the large fragments of DNA are ultrasonically fragmented. The current sample fragmentation method is the Covaris fragmentation method, which fragments the sample DNA into fragments in the range of 100-700bp. (Note: The i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com