interference plasmid pHX RNAi applied in specificity silent gene in filamentous fungi
A filamentous fungus, gene silencing technology, applied in the field of genetic engineering, to achieve good application prospects, efficient and stable silencing effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0065] (1) Preparation of protoplasts
[0066] ① Take freshly sporulated Penicillium oxalicum plate or slope, and wash the conidia with sterile saline (0.9% NaCl, 0.05% Tween) to prepare the Penicillium oxalicum conidia suspension.
[0067] ②Cover a layer of cellophane on the plate of Vogel's basic medium (2% glucose as carbon source), spread it with a glass rod, add 50-100 μl of spore suspension on the cellophane, and spread evenly. Do 8-10 similar treatments and incubate at 30°C for 12-15h.
[0068]③Add 0.1g lyase (Lysing Enzymes from Trichoderma harzianum, Sigma product number L1412) to 20ml solution 1 (solution 1: 1.2M sorbitol, 0.1M KH 2 PO 4 , pH 5.6), shake gently, and filter-sterilize with a 0.2 μm filter into a petri dish or test tube.
[0069] ④Pipe 2-3ml of lyase solution into a sterile petri dish, cover the surface of the solution with a layer of cellophane with Penicillium oxalicum bacteria, then add 2-3ml of lyase solution, and stack 6-7 layers in this order ...
Embodiment 1
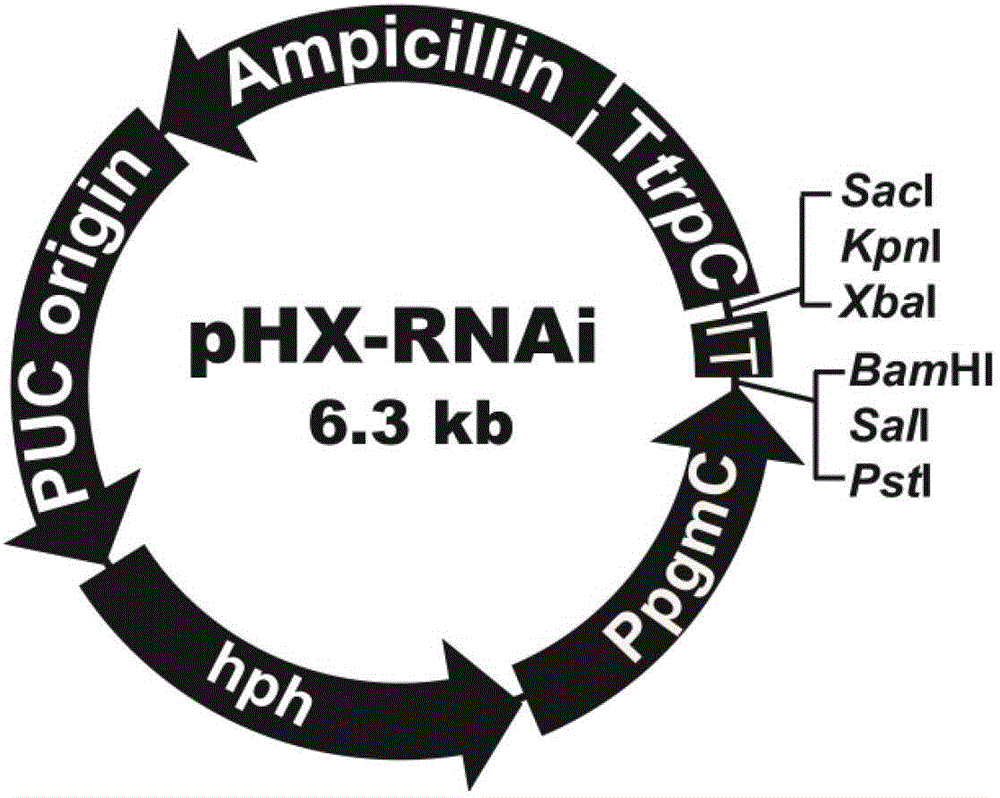
[0098] The construction of embodiment 1 interference carrier pHX-RNAi
[0099] (1) Element cloning in interference vector pHX-RNAi
[0100] Using the Penicillium oxalicum 114-2 genome as a template, PpgmC with the corresponding restriction site sequence was amplified. Using p-silent as a template to amplify hph and TtrpC, the primer sequences are as follows.
[0101] PpgmC upstream primer: Ppgm-F (SphI): AC ATGCAT GCCCGACTCATTACATACCTCCA
[0102] PpgmC downstream primer: Ppgm-R (PstI): AA CTGCAG GAGGTGGTGAAGGTTGGATTTG
[0103] TtrpC upstream primer: TtrpC-F (SacI): CG GAGCTC ACTTAACGTTACTGAAATCAT
[0104] TtrpC downstream primer: TtrpC-R (EcoRI): CG GAATTC CTAGAGCGGCCGCAACCCAG
[0105] Hph upstream primer: Hphs-F (HindⅢ): CCC AAGCTT CGACCTTAACTGATATTGAA
[0106] Hph downstream primer: Hphs-R (SphI): ACAT GCATGC CAACCCAGGGCTGGTGACGG
[0107] (2) Construction of plasmid K-hph
[0108] The hygromycin-resistant hph expression cassette hph was connected into the s...
Embodiment 2
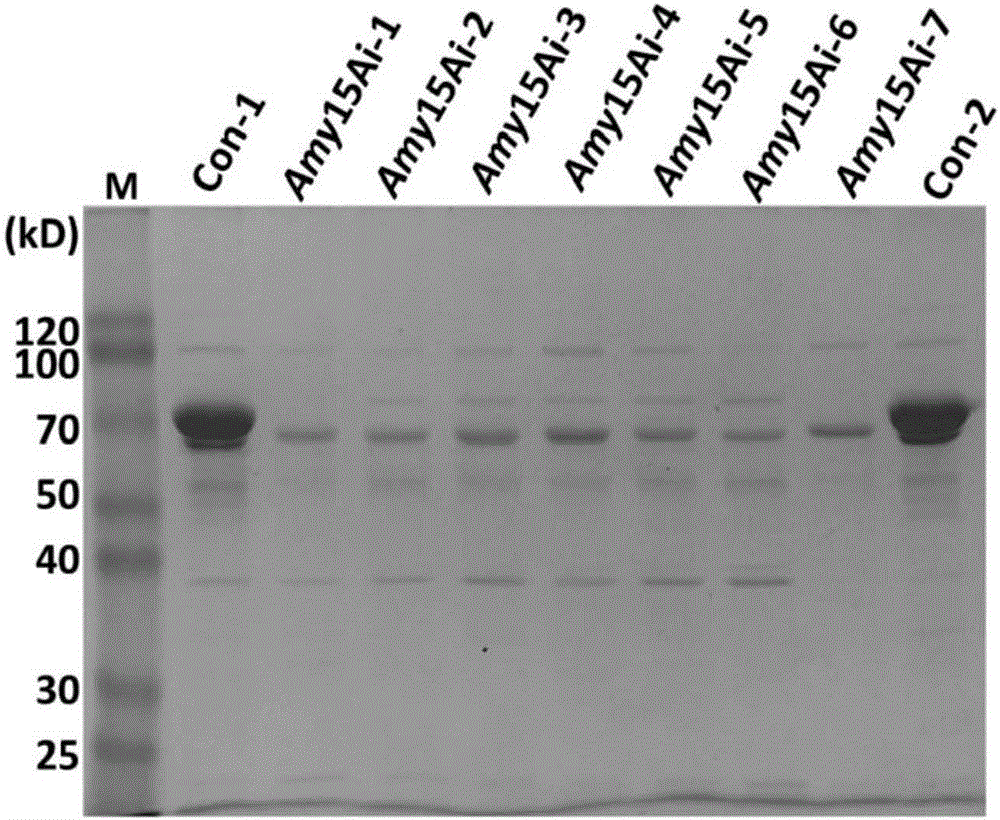
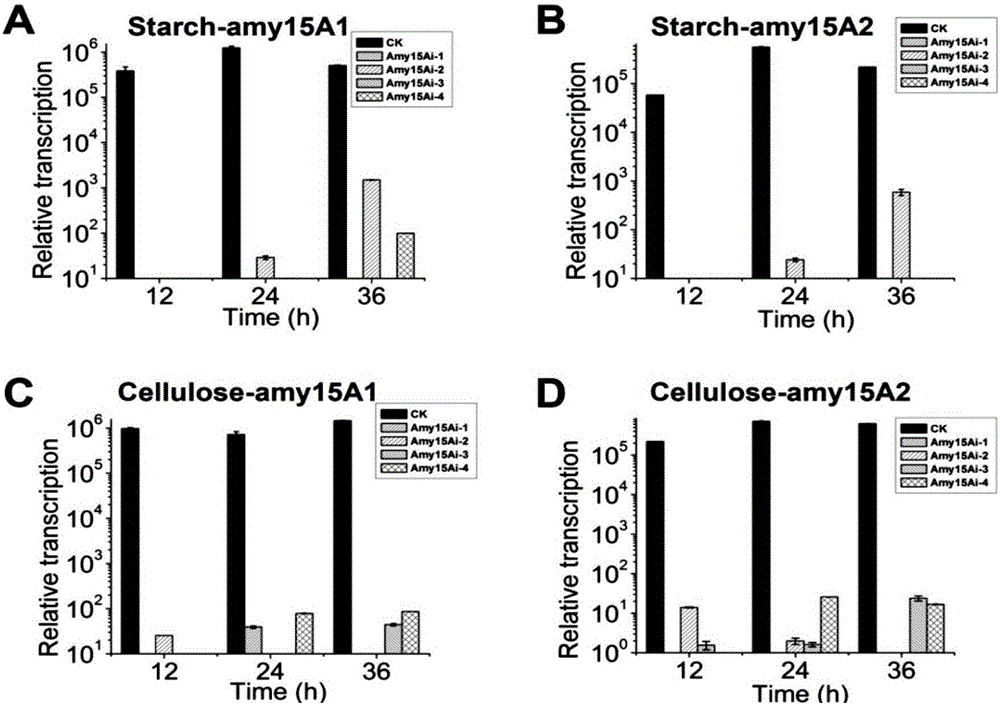
[0115] The construction of embodiment 2 single gene interference plasmid pHX-amyi
[0116] (1) Cloning of fragments
[0117] Using the 114-2 genome as a template, the amy15A sequence with corresponding restriction sites was amplified as an interference fragment, in which iamyF1(PstI) / iamyR1(BamHI) was used to amplify amy15Ai+, iamyF2(XbaI) / iamyR2(SacI) Used to amplify amy15Ai-, the primer sequences are as follows.
[0118] iamyF1(PstI): AACTGCAGTGAAAAATGACCCTCTAATGAA
[0119] iamyR1(BamHI): CGCGGATCCCCAAGCACCGGTGAAGGCG
[0120] iamyR2(SacI): CGGAGCTCCCAAGCACCGGTGAAGGCG
[0121] iamyF2(XbaI): GCTCTAGATGAAAAATGACCCTCTAATGAA
[0122] After gel electrophoresis, the PCR amplification products were cut and purified according to the instructions of the kit. The nucleotide sequences of the amy15A interference fragment are shown in SEQ ID No.2 and SEQ ID No.3.
[0123] (2) Construction of amy15A interference plasmid pHX-amyi
[0124] The amy15A-fragment and the interference plas...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com