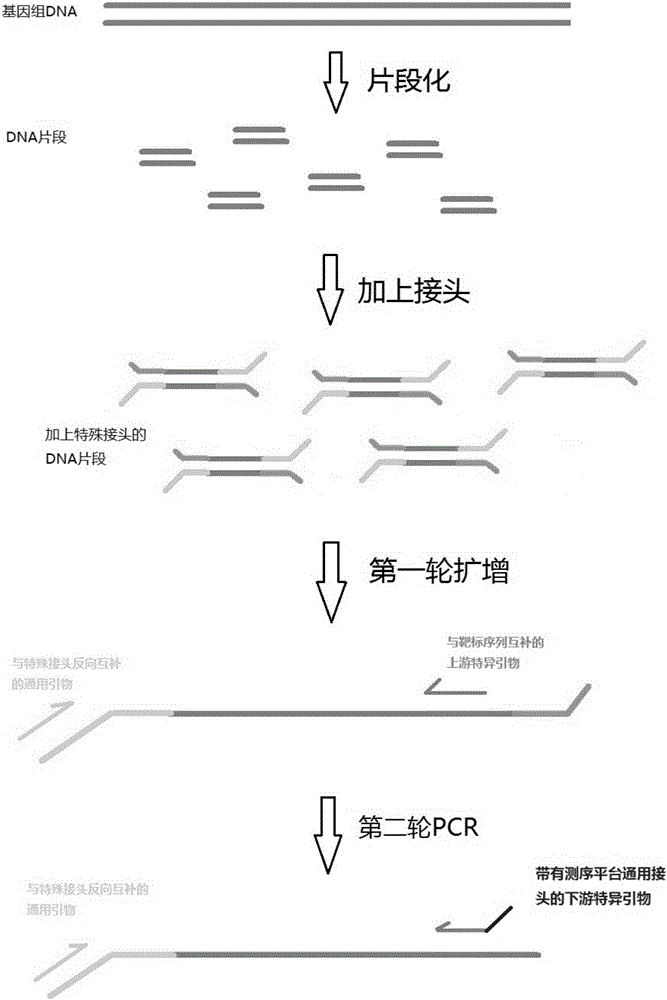
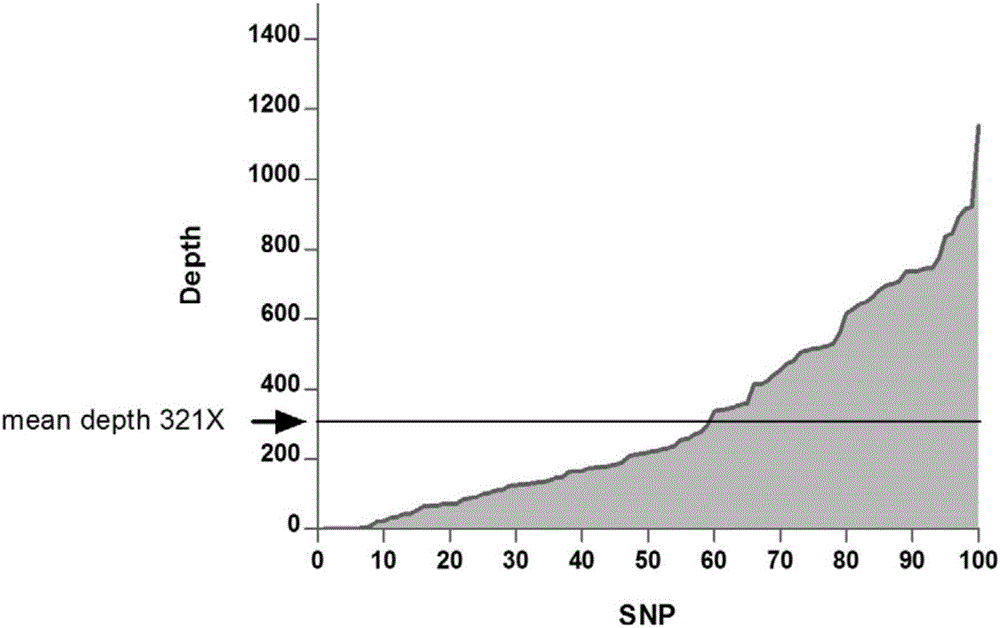
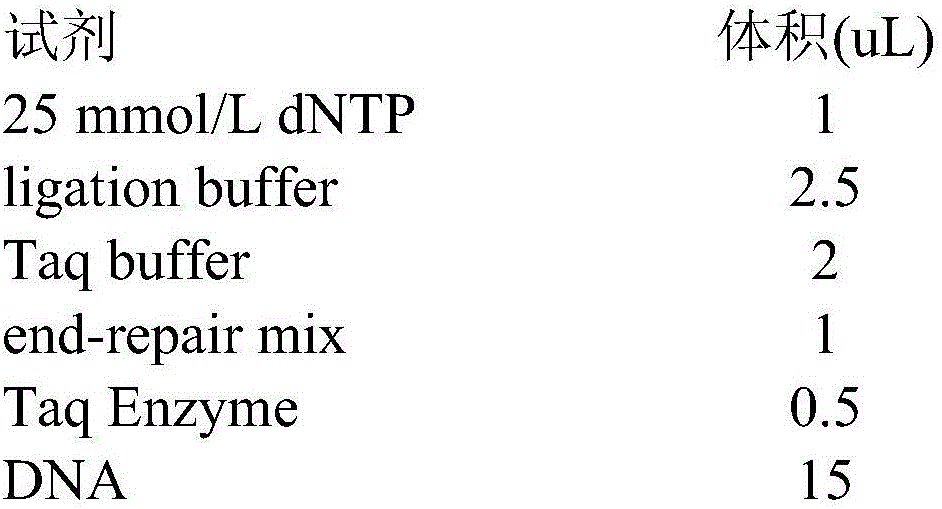Next generation sequencing library building technology based on multiplex PCR (Polymerase Chain Reaction)
A technology for a second-generation sequencing library and a construction method, which is applied in the field of second-generation sequencing library construction, can solve the problems of increased time cost, high economic cost, and reduction, and can reduce the difference in amplification efficiency, reduce the type and quantity of primers, inhibit the The effect of mutual interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1. Fragmentation of Genomic DNA
[0027] a. Turn on the Qsonica ultrasonic breaker, let the circulating water bath cool down in advance, and the working temperature is 4°C.
[0028] b. Transfer a total of 1ug of genomic DNA to a 0.2mL centrifuge tube, make up to 100uL with Nuclease Free Water (hereinafter referred to as NFW), vortex to mix, and centrifuge for a short time.
[0029] c. Put a 0.2mL centrifuge tube into the adapter of the sonicator, tighten the central shaft, insert the top cover, close the machine, and set the interruption program: amplitude -25%, on & off 20s-30 seconds, duration 15 minutes. Start interrupting.
[0030] d. After the interruption, take out the 0.2mL centrifuge tubes, arrange them neatly in order, and open the caps.
Embodiment 2
[0031] Example 2. 3in 1 connector connection
[0032] a. Configure the reaction system 22uL, first configure the reagents other than DNA, according to 125% of the theoretical value, shake and mix, centrifuge for a short time, dispense into a 96-well plate, add 7uL to each well, and then add 0.2mL from the front Transfer 15uL of DNA to each well in a centrifuge tube and cover with a cap. Vortex to mix and centrifuge briefly.
[0033]
[0034] b. Put the 96-well plate or eight tubes into the PCR machine. Setting program: 30 minutes at 25°C, 15 minutes at 72°C, hold at 4°C.
[0035] c. After finishing, add 0.5uL T4 Ligase (single gun) and 1uL Adapter to each well, and cover with a new cap. Vortex to mix and centrifuge briefly. Incubate at room temperature for 20-30 minutes.
[0036] d. Shake the magnetic beads for 30 seconds. After mixing thoroughly, add 25uL magnetic beads to each well and cover with a new cap. Vortex to mix and centrifuge briefly. Incubate for 5 minutes...
Embodiment 3
[0041] Example 3. Multiplex PCR
[0042] 1. The first round of multiplex PCR
[0043] a. Check the sample concentration according to the Qubit dsDNA HS Assay Kit operation manual. According to the sample concentration detected by Qubit, take the same amount (10ng) for each sample, mix every 5-10 samples and transfer them to a new well plate.
[0044] b. The first round of specific primers are ordinary PCR amplification primers, with a total of about 20 bases, the Tm value is set at 60°C, and the sequence is designed according to the target region. Multiple primers are mixed into an OP primer pool as the first Upstream primers for one round of PCR. Configure the total PCR system according to 125% of the theoretical value, shake and mix, and centrifuge for a short time.
[0045]
[0046] c. Put the orifice plate into the PCR instrument, and select the MAPlex-1st program to run for 2 hours.
[0047] 2. Purification of PCR products
[0048] a. After PCR, remove the well pla...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com