SNP molecular marker relating to peach tree bleeding disease resistance
A technology of peach gummosis and molecular markers, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, DNA/RNA fragments, etc., to achieve the effects of improving breeding efficiency, high accuracy, and high degree of separation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0041] The present invention will be further described below in conjunction with specific examples.
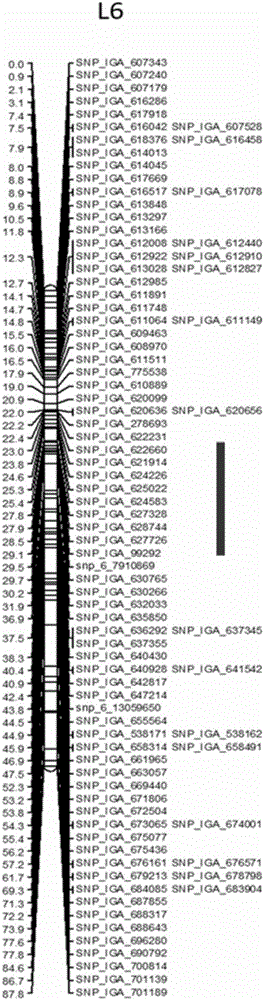
[0042] Embodiment The acquisition of the closely linked SNP molecular markers of the QTL associated with peach tree gummosis resistance comprises the following steps:
[0043] 1) Build F 1 hybrid population
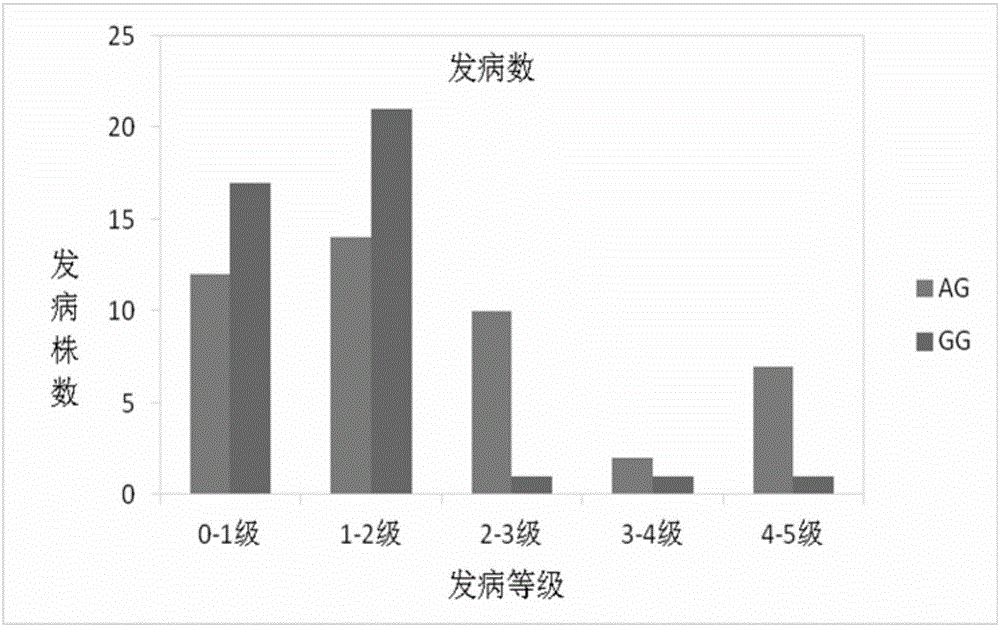
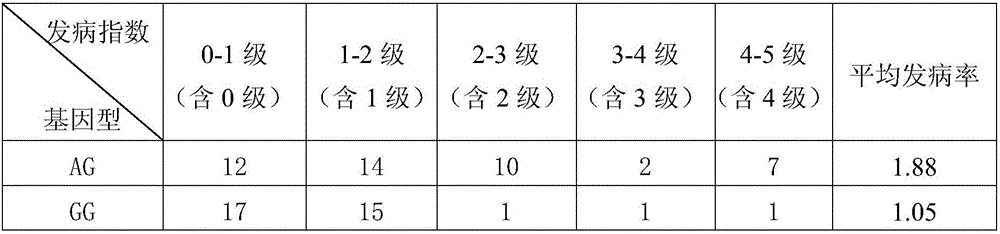
[0044] F 1 Finally, 80 hybrid offspring were obtained, and the offspring populations were segregated for gummosis traits.
[0045] 2) Genomic DNA extraction
[0046] Use the Qiagen DNeasy mini Kit kit to extract genomic DNA from 2 peach parents and 80 hybrid offspring, and follow the method provided in the kit, as follows:
[0047] ①Weigh 0.1g of frozen leaves, quickly grind them in liquid nitrogen, and place them in a 2ml sterilized centrifuge tube.
[0048] ② Add 400 μl AP1 buffer solution and 4 μl RNase A to the above centrifuge tube, vortex and mix well, bathe in 65°C water for 10 minutes, and mix up and down 2-3 times.
[0049] ③ Add 130 μl AP2 buffer solution to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com