Method for quickly modifying avidin on interface based on lipidosome
A technology of liposome and avidin, which is applied in the direction of immobilization on or in the inorganic carrier, can solve the problems of agglomeration of impurities on the glass surface, influence of single-molecule fluorescence observation, incomplete interface sealing, etc., to achieve a clean background, Uniform interface modification and time-saving effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
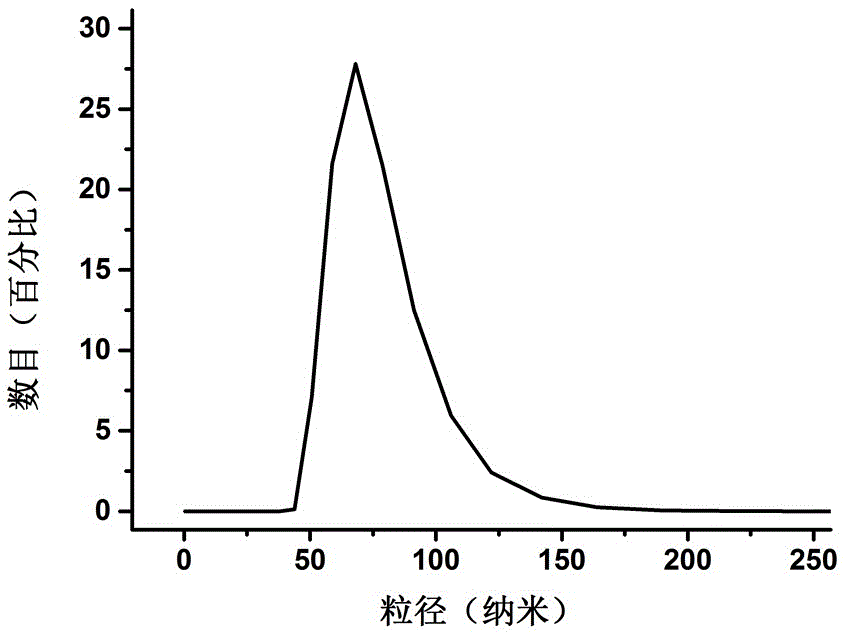
[0026] Dissolve DOPC and biotinylated phosphatidylethanol in 5 mL of chloroform at a ratio of 100:1, mix well, add flowing nitrogen to dry, and then vacuum dry for 3 hr to remove residual organic solvent. Add PBS (pH=7.4) to 1 mL of the obtained phospholipids to hydrate the monolayer phospholipids to a final concentration of 5 mg / mL. The liposomes with biotin can be obtained by extruding 20 times with an extruder with a pore size filter membrane of 50 nm. The slides were ultrasonicated with lotion and ultrapure water for 10 minutes, then ultrasonicated with acetone and sodium hydroxide solution for 30 minutes, rinsed with ultrapure water, soaked in 1% hydrofluoric acid solution for about 30 seconds, and then cleaned with ultrapure water. Rinse with pure water and blow dry with nitrogen gas for later use. A fence was attached to the glass surface, and 100 μL of the prepared liposomes were added to the fence, followed by incubation at 45°C for 10 minutes. Subsequently, the exc...
Embodiment 2
[0028] Dissolve DOPC and biotinylated phosphatidylethanolamine in 5 mL of chloroform at a ratio of 100:1, mix well, add flowing nitrogen to dry, and then vacuum dry for 3 hr to remove residual organic solvent. Add 1 mL of PBS (pH = 7.4) to the resulting phospholipids to hydrate the monolayer of phospholipids to a final concentration of 5 mg / mL. The liposomes with biotin can be obtained by extruding 20 times with an extruder with a filter membrane with a pore size of 100 nm. The slides were ultrasonicated with lotion and ultrapure water for 10 minutes, then ultrasonicated with acetone and sodium hydroxide solution for 30 minutes, rinsed with ultrapure water, soaked in 1% hydrofluoric acid solution for about 30 seconds, and then cleaned with ultrapure water. Rinse with pure water and blow dry with nitrogen gas for later use. A fence was attached to the glass surface, and 100 μL of the prepared liposomes were added to the fence, followed by incubation at 45°C for 10 minutes. Su...
Embodiment 3
[0030] Dissolve DOPC and biotinylated phosphatidylethanolamine in 5 mL of chloroform at a ratio of 100:1, mix well, add flowing nitrogen to dry, and then vacuum dry for 3 hr to remove residual organic solvent. Add PBS (pH = 7.4) to the resulting phospholipids to hydrate the monolayer phospholipids to a final concentration of 5 mg / mL. The liposomes with biotin can be obtained by extruding 20 times with an extruder with a 200 nm pore size filter. The slides were ultrasonicated with lotion and ultrapure water for 10 minutes, then ultrasonicated with acetone and sodium hydroxide solution for 30 minutes, rinsed with ultrapure water, soaked in 1% hydrofluoric acid solution for about 30 seconds, and then cleaned with ultrapure water. Rinse with pure water and blow dry with nitrogen gas for later use. A fence was attached to the glass surface, and 100 μL of the prepared liposomes were added to the fence, followed by incubation at 45°C for 10 minutes. Subsequently, the excess liposom...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com