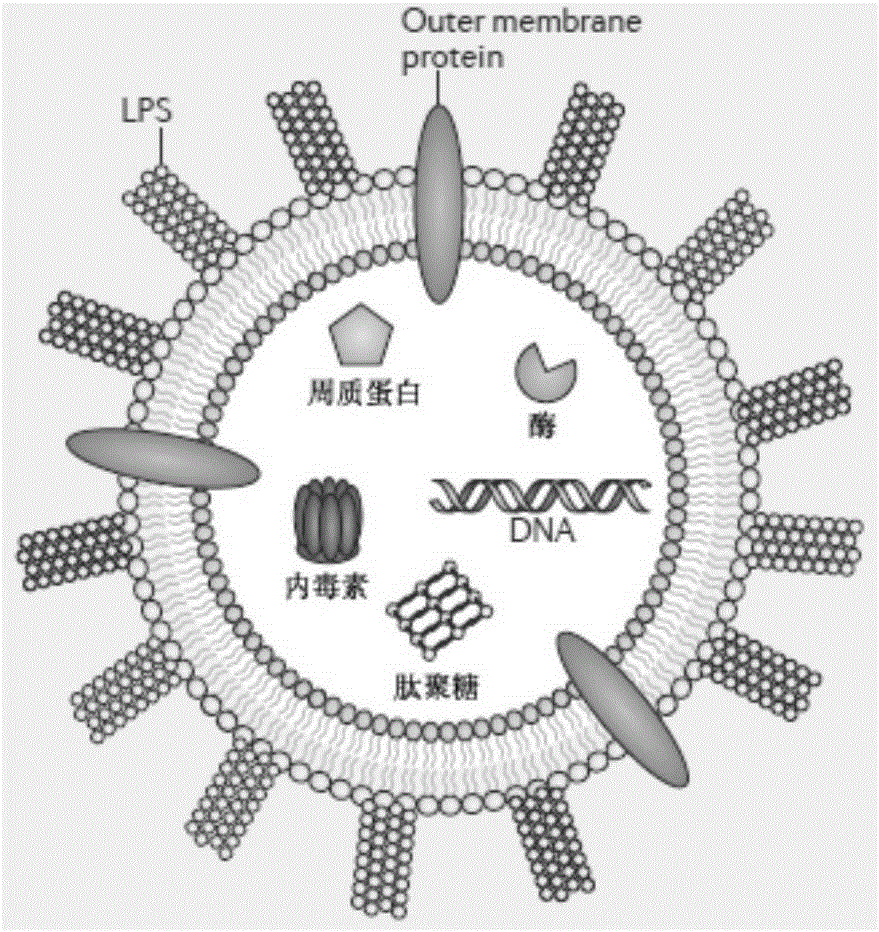
Method for extracting bacterial outer membrane vesicles
A technology for outer membrane vesicles and membrane vesicles, which is applied in the field of preparing Escherichia coli and Klebsiella pneumoniae efflux membrane vesicles, can solve problems such as troublesome handling and complicated operation, and achieve the effect of convenient operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1: Selection and cultivation of bacterial strains
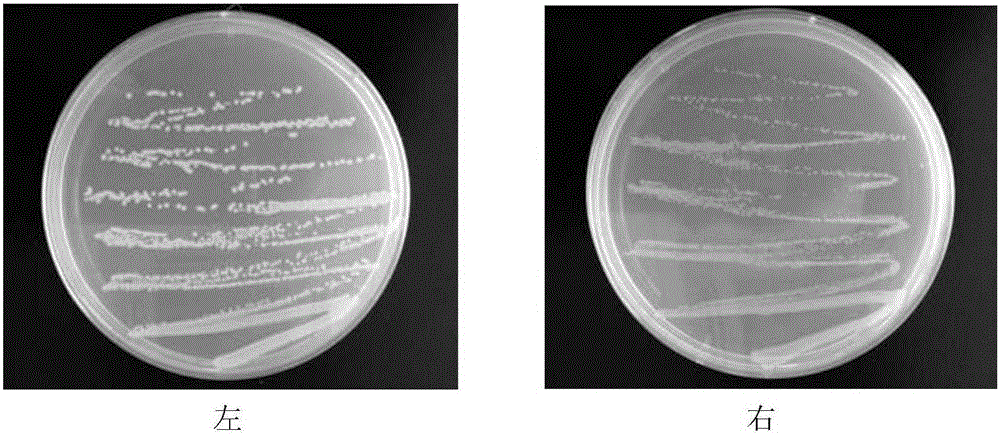
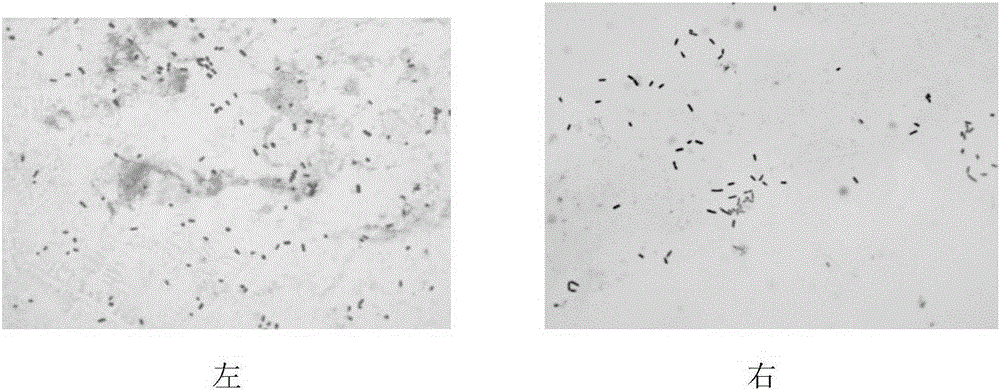
[0027] Klebsiella pneumoniae was purchased from the China Agricultural Microorganism Culture Collection, and Escherichia coli DH5α was purchased from the American Culture Collection. Klebsiella pneumoniae was cultured with NB medium, and Escherichia coli was cultured with LB medium; the freeze-dried powder of the strains was dissolved in sterile saline, and an appropriate amount was placed on a solid medium, and cultured overnight at 37°C until a single colony (such as figure 2 shown); single colonies were cultured in test tubes containing 3ml of liquid medium, and cultured overnight at 37°C on a shaker at 180r / min. The next day, transfer 3ml of the bacterial solution to a shake flask containing 40ml of medium for cultivation until the OD value of the bacteria reaches 1.0. image 3 The cell morphology after crystal violet staining is shown.
Embodiment 2
[0028] Embodiment 2: Extraction of OMVs
[0029] Centrifuge the bacterial solution cultivated to an OD value of 1.0 in a 50ml sterile centrifuge tube at 6000r / min for 20min, remove most of the bacterial cells, filter the supernatant with a 0.22μM sterile filter head to completely sterilize, and then transfer the filtrate Centrifuge at 3000r / min for 10min at 4°C for 10min in 50ml of ultrafiltration tube with a molecular weight cut-off of 100KD, and repeat several times until the solution is overall and concentrated to about 1 / 8 of its original volume;
[0030] Add 5ml of normal saline to the concentrated solution, use the ultrafiltration concentration method to wash away impurities such as excess protein and flagella pili, dispense it into EP tubes, and store at -20°C.
Embodiment 3
[0031] Example 3: Identification of OMVs
[0032] A, BCA method to the determination of OMVs protein concentration;
[0033] OMVs protein concentration was measured by BCA method;
[0034] Take 5 mg / ml standard protein and establish protein-absorbance standard curve in 96-well plate (such as Figure 4 As shown), take 2 μl OMVs samples in the other 5 auxiliary wells, add 200 μl AB solution (A solution: B solution = 50:1) in each well, incubate in a 37°C incubator for 30min, measure the absorbance value of each well at a wavelength of 570nm ; To establish a standard curve for protein content, R 2 =0.9989>0.99, showing a good linear relationship;
[0035] The measured protein concentration of Klebsiella pneumoniae OMVs is about 1.5 mg / ml, and the concentration of Escherichia coli OMVs is about 2.1 mg / ml;
[0036] Table 1 shows the protein concentration of the extracted Klebsiella pneumoniae and Escherichia coli OMVs.
[0037] Table 1
[0038]
[0039] B. Negative stain ele...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com