LAMP primer set for quickly detecting salmonella and application thereof
A Salmonella and primer set technology, applied in the biological field, can solve the problems of insufficient versatility and specificity of primers, inability to amplify nucleic acids, primer mismatches, etc., and achieve high repeatability, efficient amplification, and primer specificity. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] This example is used to illustrate the versatility of the primer set of the present invention for the detection of different forms of LAMP. The different forms of LAMP detection include electrophoresis LAMP, precipitation LAMP and chromogenic LAMP. The steps are as follows:
[0047] (1) Concentration gradient dilution of Salmonella
[0048] The Salmonella strain used for detection comes from the China Industrial Microbiology Culture Collection and Management Center, and the number is CICC10420 Salmonella typhimurium. In a liquid medium, culture overnight at 36°C, centrifuge the pure culture of Salmonella at 12000rpm for 10min, discard the supernatant, wash the precipitate with 0.9% normal saline and centrifuge once, and wash the precipitate with ddH 2 O resuspended, and the concentration of bacteria was calibrated by McFarland turbidity method, that is, 1MCF≈3×10 8 cfu / mL. were diluted to 10 6 cfu / mL, 10 5 cfu / mL, 10 4 cfu / mL, 10 3 cfu / mL, 10 2 cfu / mL, 10 1 cfu / ...
Embodiment 2-5
[0060] Embodiment 2-5 Salmonella reaction system and detection method
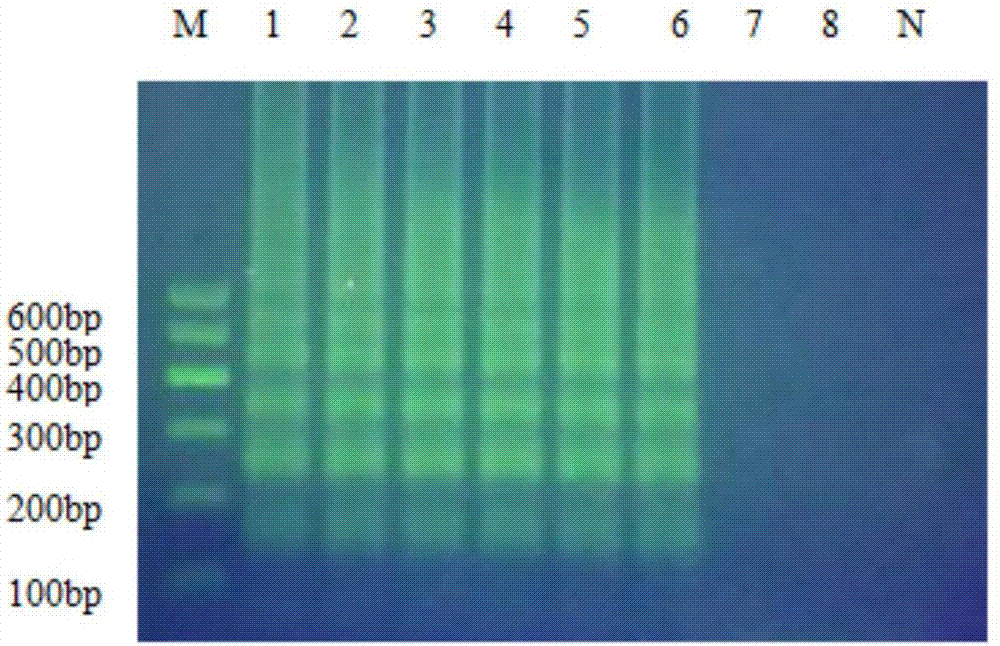
[0061] The concentration gradient dilution of Salmonella and the extraction steps of genomic DNA are as steps (1) and (2) in Example 1, and the reaction system is shown in Table 3, wherein the primers of Examples 2 and 3 adopt primer set A, and the primers of Examples 4 and 3 Primer set B was used for primer 5, and the amplification results were confirmed by turbidity detection. It can be seen from Table 3 that the primer set, detection method and reaction system of the present invention can well amplify the Salmonella specific fragment and obtain detection results.
[0062] Each reaction system, reaction condition and reaction result of table 3 embodiment 2~5
[0063]
[0064]
Embodiment 6
[0066] This example is used to illustrate the versatility of the primer set of the present invention to Salmonella.
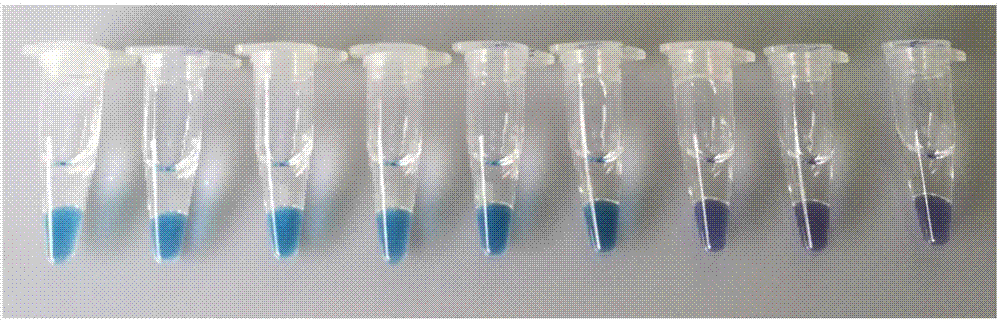
[0067] According to the method in Example 1, DNA was extracted from 5 kinds of Salmonella (Table 4), amplified by LAMP, and detected by chromogenic method LAMP.
[0068] The results of the experimental group are as Figure 4 As shown, the amplification reactions of the Salmonella strains of the A and B primer groups were all positive, showing blue-green, and the reaction tube of the negative control (NTC) was blue-purple without change. It shows that the primer set of the present invention has better versatility to the above five kinds of Salmonella.
[0069] Table 4 primer group of the present invention is used bacterial strain and detection result to the universality test of Salmonella
[0070]
[0071] Note: 1. CMCC: China Medical Bacteria Culture Collection Management Center; CICC: China Industrial Microbiology Culture Collection Management Center. 2....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com