Recombinant pichia pastoris for enhanced expression of xylanase from Streptomyces sp.FA1
A technology of Pichia pastoris and xylanase, applied in the field of genetic engineering, can solve the problems such as episomal expression plasmids that have not yet been seen, and achieve the effect of improving specific activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
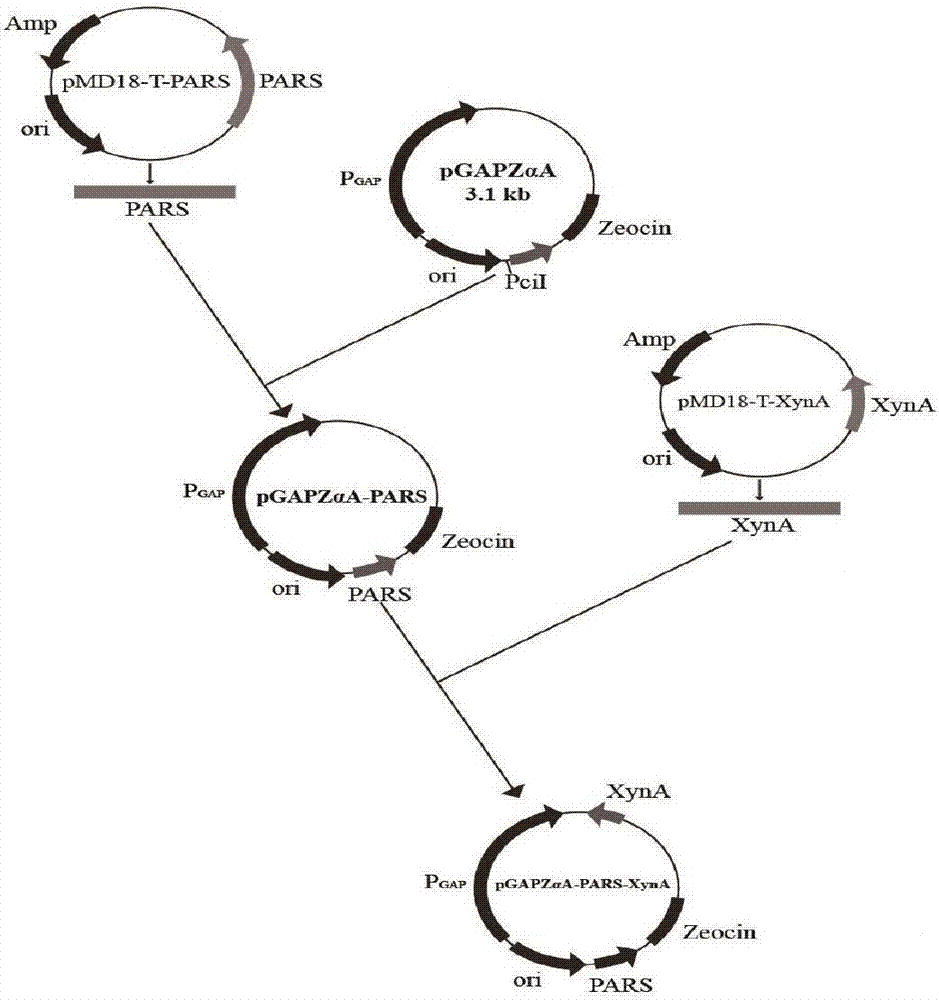
[0034] Example 1: Construction of expression vector pGAPZαA-PARS
[0035] The self-replicating sequence PARS with nucleotide sequence as shown in SEQ ID NO.2 was synthesized by whole gene synthesis technology; the recovered PARS gene fragment was connected with plasmid pGAPZαA by infusion enzyme to construct the expression vector pGAPZαA-PARS.
Embodiment 2
[0036] Example 2: Production of Pichia pastoris KM71 / pPIC9K-XynA Competent Cells:
[0037]1) Pick a single colony of yeast KM71 / pPIC9K-XynA, inoculate it into a 50ml Erlenmeyer flask containing 10ml of YPD medium, and culture overnight at 30°C and 250rpm;
[0038] 2) Inoculate 100 μL of cultured cells into a 500ml Erlenmeyer flask containing 100mL of LYPD medium, culture overnight at 30°C and 250rpm until OD600 reaches 1.3-1.5;
[0039] 3) Centrifuge three 50ml sterile centrifuge tubes at 4°C, 5000rpm for 5min, discard the supernatant, add 4ml sterile water to each tube to fully suspend the cells, collect and combine them into one tube; add 2ml 10*TE buffer solution (PH=7.5), 2ml 10 *LiAC and 0.5ml 1M DTT were rotatably mixed, and shaken in a water bath at 30°C and 50rpm for 45min.
[0040] 4) Dilute the bacterial suspension in 3) with sterile water to 30ml, centrifuge at 5000rpm at 4°C for 5min, discard the supernatant to collect the bacterial cells, and add 25mL of pre-cool...
Embodiment 3
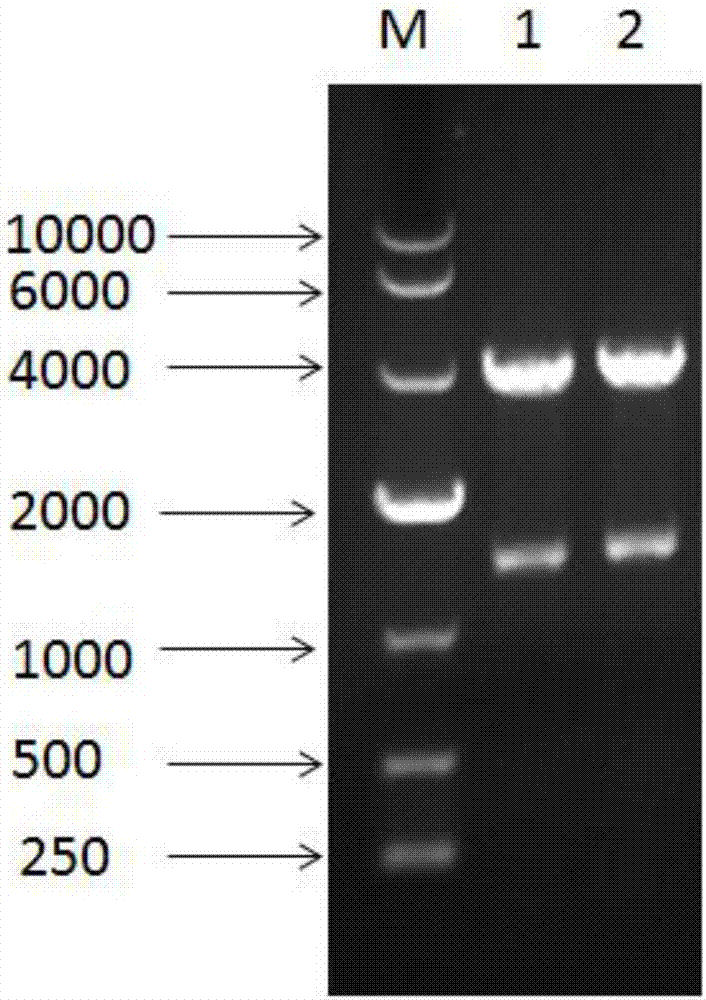
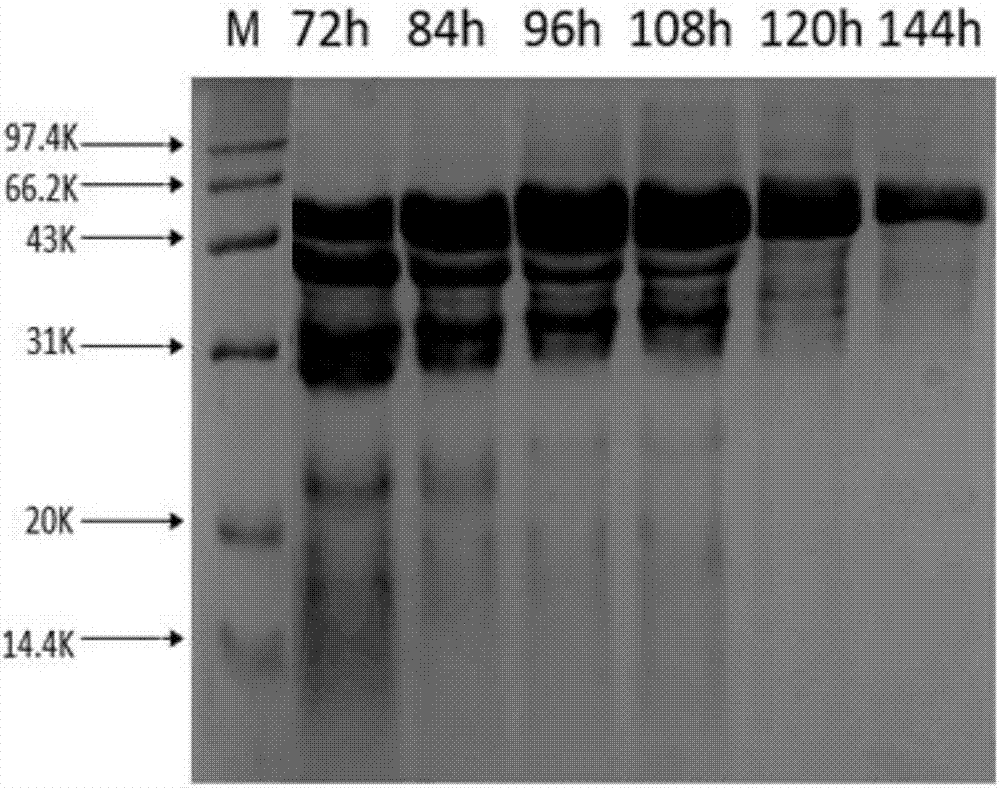
[0044] Example 3: Construction of Enhanced Expression Recombinant Bacteria KM71 / pPIC9K-XynA / pGAPZαA-PARS-XynA
[0045] Construction of recombinant plasmid pGAPZαA-PARS-XynA:
[0046] The plasmid pMD18-T-XynA (A xylanase from Streptomyces sp.FA1: heterologous expression, characterization, and its application in Chinese steamed bread, YangXu, Journal of Industrial Microbiology & Biotechnology, May 2016, Volume43, Issue5, pp 663-670) was used as the template A forward primer with the sequence shown in SEQ ID NO.3: CCGGAATTCATGGCCGAGAACACCCTT and a reverse primer with the sequence shown in SEQ ID NO.4: ATTTGCGGCCGCTCAGGTGCGGGTCCAGCGTT were designed. The XynA fragment with Not I and EcoR I restriction sites was obtained by polymerase chain reaction.
[0047] PCR reaction system (50μL):
[0048]
[0049] PCR program: 94°C, 4min (pre-denaturation); 98°C, 10s (denaturation); 60°C, 5s (annealing); 72°C, 90s (extension); set 30 cycles; set after 72°C, 10min (insulation) The storag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com