SNP functional molecular marker of lodging-resistant gene of japonica rice and application thereof
A functional molecule and anti-lodging technology, applied in the field of plant molecular biology, can solve the problems of long time-consuming identification of lodging resistance phenotypes, inaccurate manual identification results, and unsafe experimental operations, so as to achieve accurate and reliable detection results and shorten the breeding cycle , the effect of a wide range of application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
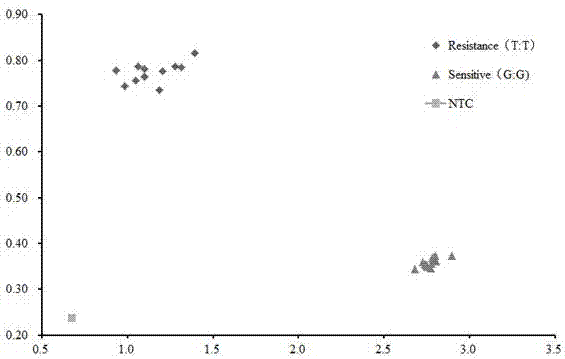
[0033] Example 1: Phenotypic verification of natural genetic material of SNP marker K_06g_SCM2-1
[0034] 23 natural genetic materials were selected for verification, including 11 lodging-resistant materials and 12 lodging-sensitive materials.
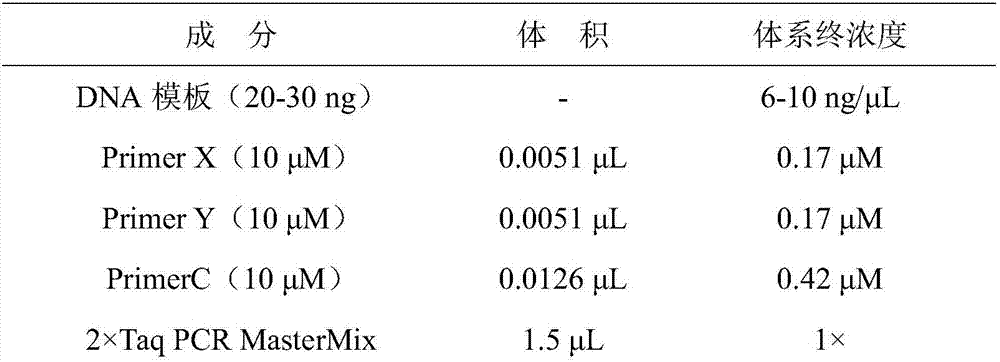
[0035] 1. Genomic DNA was extracted from japonica rice leaves by simplified CTAB method.
[0036] (1) Freeze-dry or dry the japonica rice leaves, put an appropriate amount of leaves into a 2.0mL centrifuge tube, add two steel balls, and grind them on a tissue grinder;
[0037] (2) Add 750 μL of CTAB solution, oscillate to homogenate, and incubate at 65°C for 0.5-1 h;
[0038] (3) Cool to room temperature, add 750 μL of chloroform:isoamyl alcohol (24:1) in the fume hood and mix by inverting 3-4 times;
[0039] (4) Centrifuge at 12000rmp for 10min, transfer 500μL supernatant to a new 1.5mL centrifuge tube;
[0040] (5) Add an equal volume of isopropanol solution and shake gently to mix, precipitate at -20°C for more than 1 hour, centr...
Embodiment 2
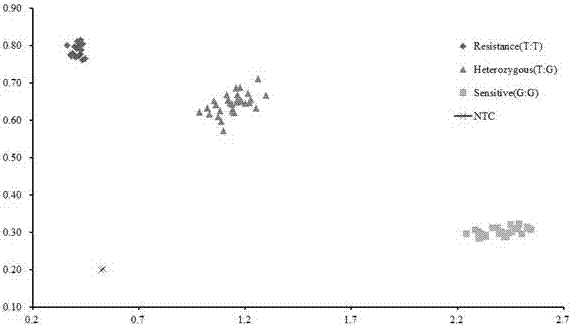
[0055] Example 2 Phenotype verification of F2 segregation population of SNP marker K_06g_SCM2-1
[0056] Detect the individual plant genotype of the F2 segregation population crossed by the donor parent Habataki and the recipient parent Sasanishiki;
[0057] 1. The genotyping method is the same as in Example 1. For the genotyping results of 80 F2 isolated populations, see figure 2 .
[0058] 2. Select two homozygous genotypes for phenotypic identification. During the period from the booting stage to the heading of rice, remove the top leaves of the plants, measure the mechanical strength and diameter of the plant stems, and comprehensively evaluate the lodging resistance of the plants.
[0059] The genotype and phenotype data of homozygous individual plants are shown in Table 3.
[0060] Table 3 The phenotype identification results of the F2 segregation population of marker K_06g_SCM2-1
[0061]
[0062]
[0063] Note: "R" indicates that the plant phenotype is resist...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com