Reproduction marker gene vasa of Sinopotamon henanense and application thereof
A technology for marking genes and genes, applied in the application, genetic engineering, plant genetic improvement and other directions, can solve the problem of gene vasa separation that still needs to be further studied
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1. Cloning and isolation of Henan Huaxi crab gene vasa.
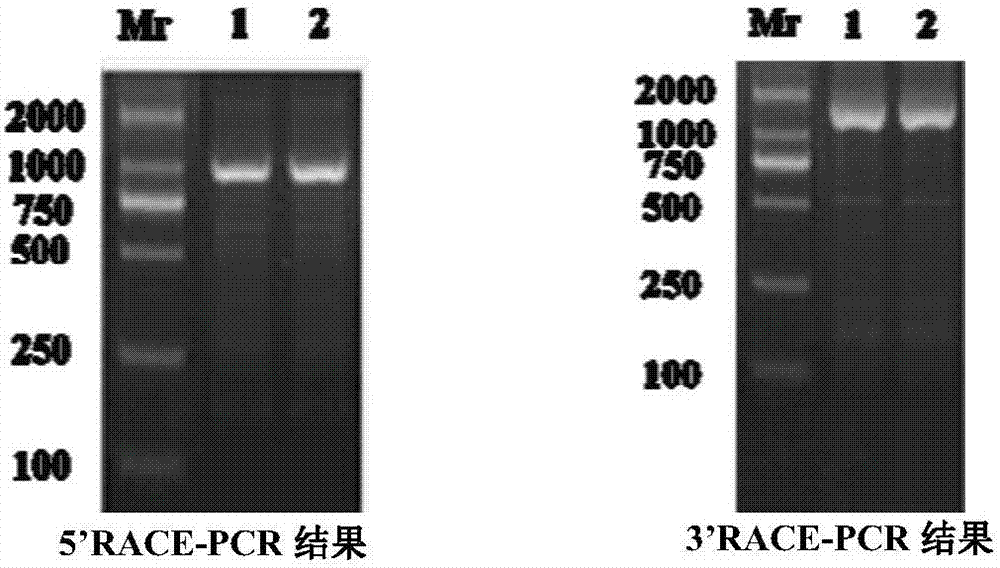
[0027] 1.1 Cloning of the partially conserved sequence of Huaxi crab vasa in Henan.
[0028] 1.1.1 Extraction of total RNA from gonad tissue and synthesis of cDNA first strand.
[0029] According to the operating instructions of the RNeasy Mini Kit (Qiagen) kit, total RNA was extracted from the ovaries or testes of early-stage Huaxi crabs in Henan. The integrity of the total RNA was detected by 1% agarose gel electrophoresis, and the concentration and purity of the total RNA were detected by a BioSpectrometer micro-spectrophotometer. According to the instructions of the TransScript One-Step gDNA Removal and cDNASynthesis SuperMix (full gold) kit, the first strand of cDNA was synthesized.
[0030] 1.1.2 Cloning of partially conserved sequences of vasa.
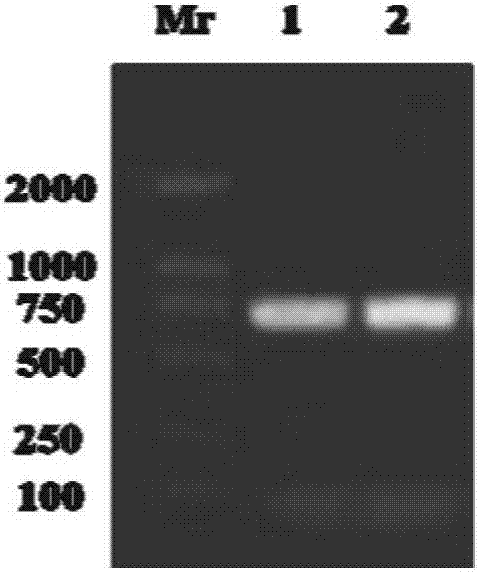
[0031] According to the conserved sequence of VASA protein in crustaceans reported in the public database, search for its corresponding nucleotide sequ...
Embodiment 2
[0058] Example 2. Structural analysis of the vasa gene sequence and VASA protein sequence of Henan Huaxi crab.
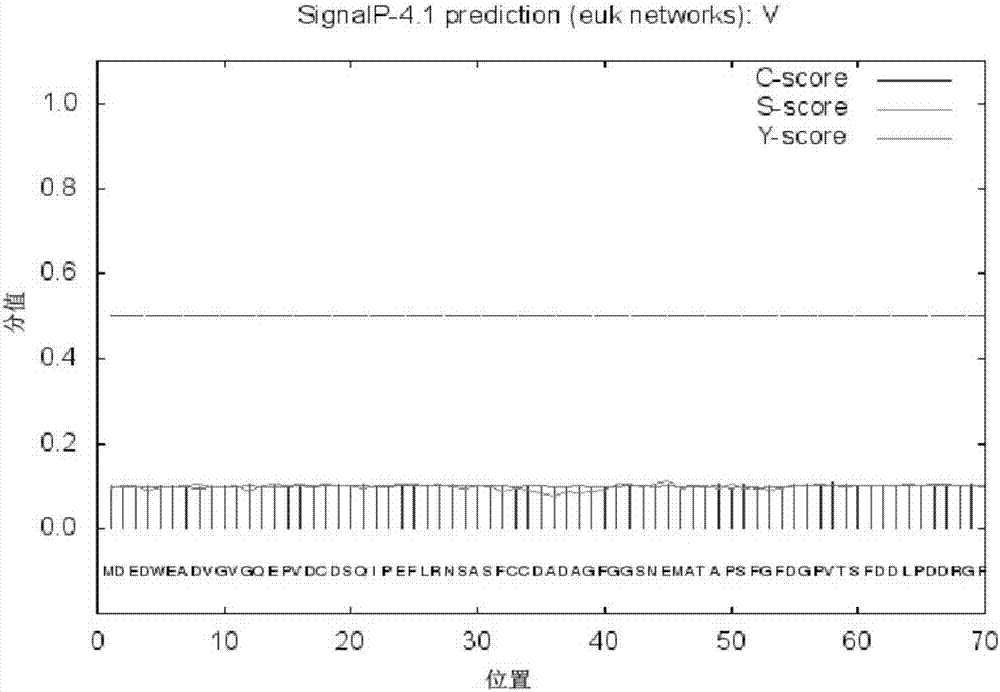
[0059] Analyze and compare the obtained vasa nucleotide sequences, and use NCBI / BLASTn (http: / / www.ncbi.nlm.nih.gov / BLAST) for homologous alignment; use NCBI / ORF Finder (http: / / www.ncbi.nlm.nih.gov / projects / gorf / orfig.cgi) and Expasy-Translate tool (http: / / web.expasy.org / translate / ) online tools to analyze and predict gene coding regions and their encoded proteins Sequence (Sequence Listing SEQ ID NO. 2).
[0060] Analyze predicted protein sequences and structures. The amino acid composition and basic physicochemical properties of SHVASA protein were analyzed using Expasy (http: / / web.expasy.org / protparam / ) online software. The results showed that the molecular weight of the SHVASA protein was 75247.8KD; the theoretical isoelectric point was 5.00, and it contained 101 negatively charged amino acid residues (aspartic acid and glutamic acid), and 76 positively charg...
Embodiment 3
[0061] Example 3. RT-PCR detection of the specific expression of the gene vasa in the gonad tissue of Henan Huaxi crab.
[0062] According to the Henan Huaxi crab vasa full-length sequence obtained in 1.3 of Example 1, referring to the results of sequence analysis in Example 2, the species-specific segment in the Henan Huaxi crab vasa sequence was selected, and the gene-specific gene used for quantitative PCR was designed. With primers, rpl38 gene (60S ribosomal protein L38) was used as an internal reference gene, and the expression of gene vasa in different tissues of Henan Huaxi crab was detected by quantitative PCR.
[0063] Using the method described in 1.1.1 of Example 1, extract total RNA from tissues such as Henan Huaxi crab ovary, testis, accessory gonads, gills, heart, hepatopancreas, intestine, stomach, muscle, etc.; synthesize the first strand by reverse transcription cDNA; use Vq and R38 as primers respectively, as shown in the sequence table SEQ ID NO.9-12; use th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com