Method for capturing cells or biomolecules in solutions
A technology of biomolecules and cells, applied in the methods of supporting/immobilizing microorganisms, cell dissociation methods, biochemical equipment and methods, etc., which can solve the problems of low specificity, inability to collect and re-cultivate circulating tumor cells, and rely on old-fashioned chips.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
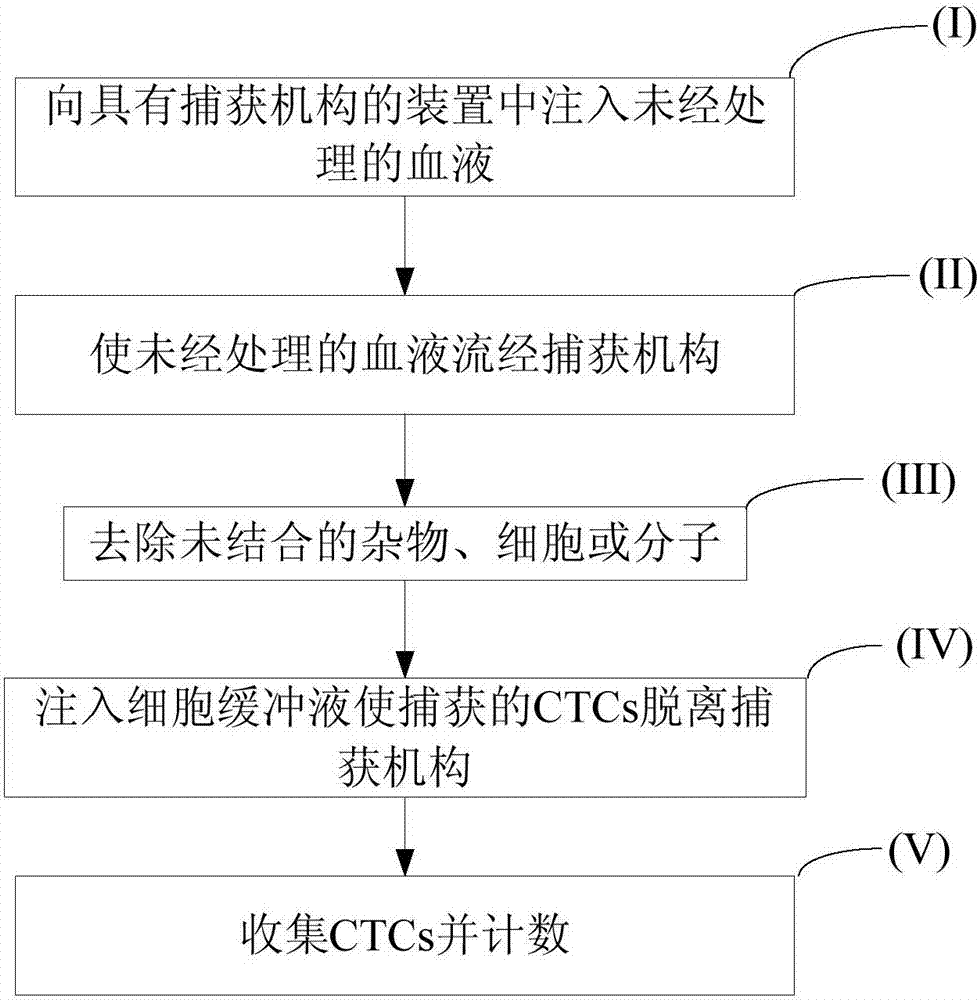
[0080] Herein, a preparation method of the capture sieve for sorting CTCs is presented. The preparation method includes:
[0081] (I) selection of sieves;
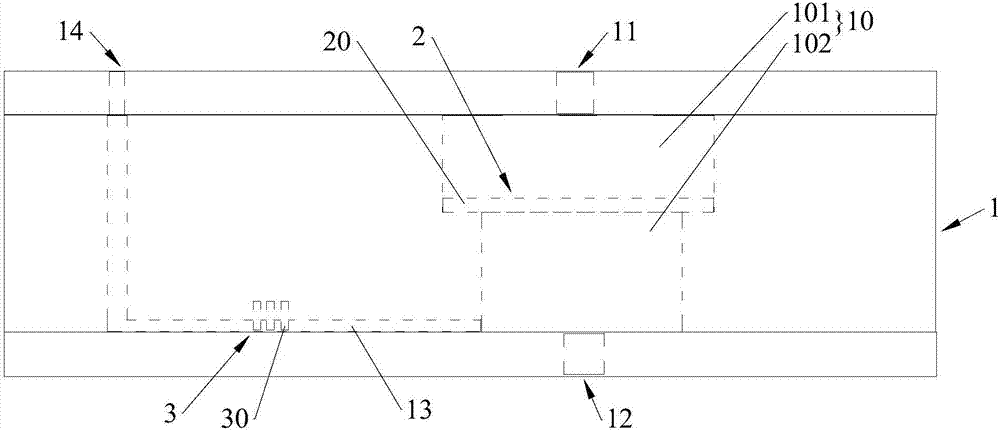
[0082] Gold sieve: Choose a gold sieve with pores (eg, 64 µm or 40 µm) to maximize contact time with tumor cells while preventing the risk of clogging. The size of the gold sieve used in the following tests is 2×2mm 2 . Larger sizes are preferred depending on the specific operation.
[0083] Stainless steel and gold sieves: 51µm pores are selected and AuPd is coated by magnetron sputtering. Such sieves are cheaper and mechanically stronger than gold, so they are more suitable for integration in devices.
[0084] (II) Pre-functionalization;
[0085] Various cleaning methods were used to prepare the sieves prior to functionalization, including autoclaving, oxygen plasma cleaning, and ultrasonic cleaning in various solutions, including piranha solutions. For example, the best results for a 64 µm gold sieve included son...
example 2
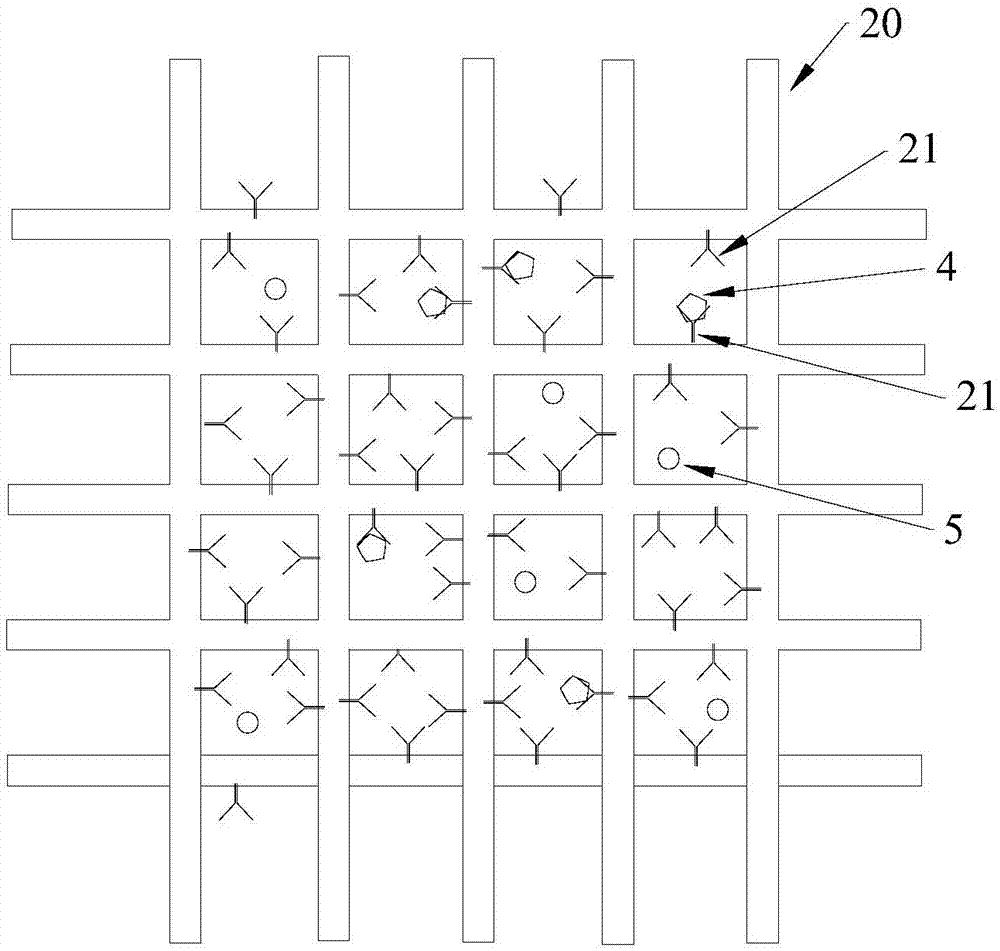
[0095] Experiments were performed to demonstrate the efficiency of the screen in capturing EpCAM expressed by tumor cells. In this case, the mesh opening is 51 μm.
[0096] Cell Selection - Cells with high expression levels of EpCAM protein (such as CaCo2 and MCF7 cells) were used.
[0097] Cell Growth - Grow in DMEM buffer at 37°C.
[0098] Cell preparation and incubation of screens - CaCo2 and MCF7 cells differentiated 1:2. The test was performed on a rotating 37°C hot plate. After isolation, cells were diluted 1:10 in DMEM buffer, and 0.5 ml was used to incubate the capture screen for 1 hour.
[0099] For washing of non-specifically bound cells, the sieves were rinsed with pure water, incubated in pure aqueous solution for 2 minutes, and then rinsed again before microscopic observation.
[0100] Microscopic evaluation of trapped cells: photograph of the sieve taken by the microscope ( Figure 5 a and 5b). Figure 5 a shows CaCo2 cells expressing the capture molecule o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com