OsbHLH116 gene for controlling rice seed germination, RNAi carrier, as well as preparation method and application thereof
An osbhlh116 and rice seed technology, applied in the field of plant genetic engineering, can solve the problems of high cost, cumbersome operation, long test period and the like, and achieve the effects of high cost efficiency, simple operation and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
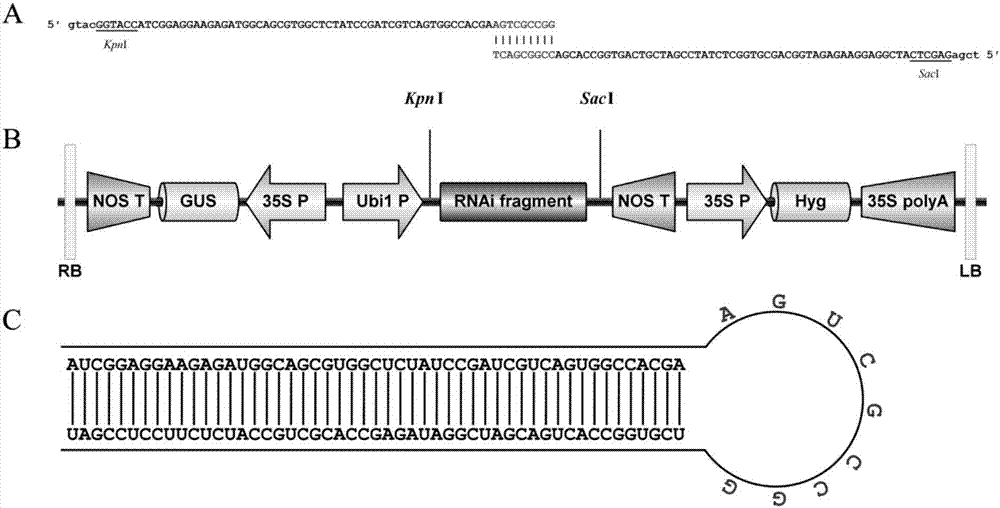
[0039] Design and Synthesis of OsbHLH116 Interfering Fragments
[0040] According to the principle of simple RNAi technology (Higuchi et al., 2009; Liu Yanxia et al., 2014), a 60bp sequence (5′-ATCGGAGGAAGAGATGGCAGCGTGGCTCTATCCGATCGTCAGTGGCCACGAAGTCGCCGG-3′, such as SEQ ID NO. 3) as a small interference fragment (positive primer OsbHLH116-F), it was compared in the rice genome database (http: / / rapdb.dna.affrc.go.jp / ), and no homologous sequence was found. The 9 bases at the 3' end (5'-AGTCGCCGG-3') and the reverse complement (5'-CCGGCGACT-3') were used as the 9 bases at the 3' end of the reverse primer (OsbHLH116-R), and the remaining 51 bases base as the 5'-end sequence of the reverse primer (e.g. figure 1 A), the primer sequence for synthesizing the RNA interference fragment of OsbHLH116 gene: siRNA-F: 5′-gtac GGTACC ATCGGAGGAAGAGATGGCAGCGTGGCTCTA TCCGATCGTC AGTGGCCACG AAGTCGCCGG-3' (as shown in SEQ ID NO. 4); siRNA-R: 5'-tcga GAGCTC ATCGGAGGAA GAGATGGCAG CGTGGCTCTA T...
Embodiment 2
[0043] Construction of OsbHLH116-RNAi expression vector
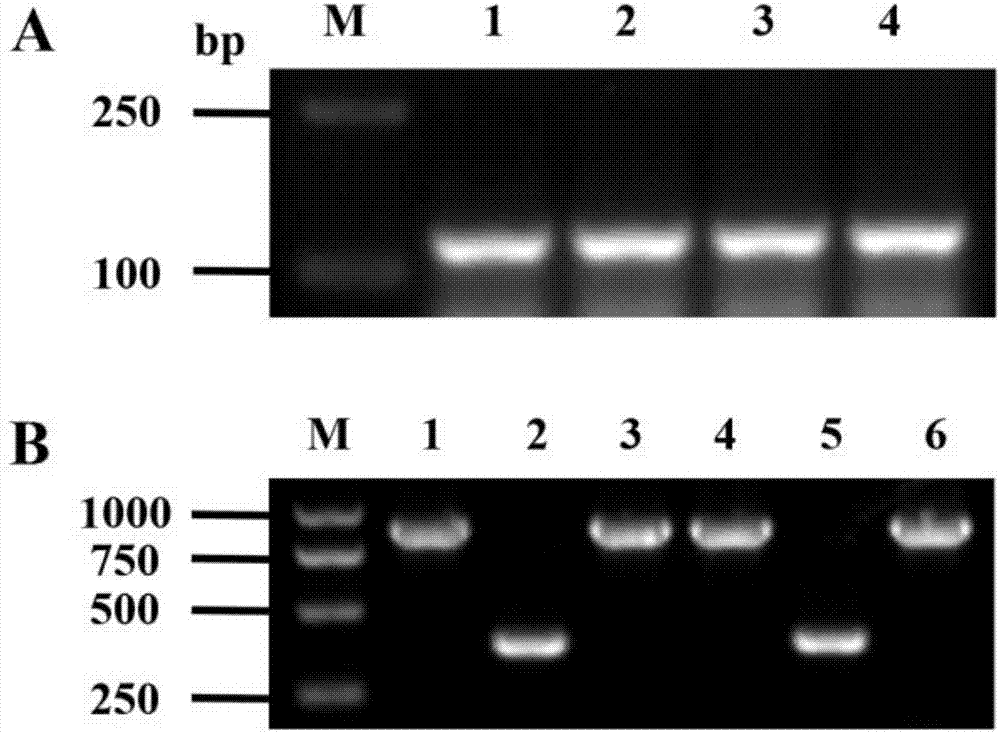
[0044] Use restriction enzymes KpnI and SacI to digest pTCK303 plasmid and OsbHLH116 small interference fragment respectively. The 50 μL digestion system is as follows: pTCK303 plasmid 2 μg (OsbHLH116 small interference fragment 1 μg), 10×L Buffer 5 μL, KpnI10U, SacI 10U, add ddH 2 O to 50μL, digested at 37°C for 4h. The digested products were purified and added to the following ligation system: 4 μL of linear plasmid, 4 μL of small interference fragment, 1 μL of 10×Buffer, 1 μL of T4 DNA ligase, and ligated overnight at 4°C. The ligation product was transformed into Escherichia coli DH5α competent cells by heat shock method, and the bacterial solution was coated on a flat plate containing 50 mg / L kanamycin, and after overnight culture, a monoclonal shaking bacterium was selected for propagation, and primer pTCK303-VF (such as SEQ ID NO. 6) and pTCK303-VR (shown in SEQ ID NO. 7) for colony PCR validation. The size of ...
Embodiment 3
[0046] Application of RNAi vectors in rice breeding.
[0047] Agrobacterium-mediated transformation of rice callus and detection of positive transgenic plants
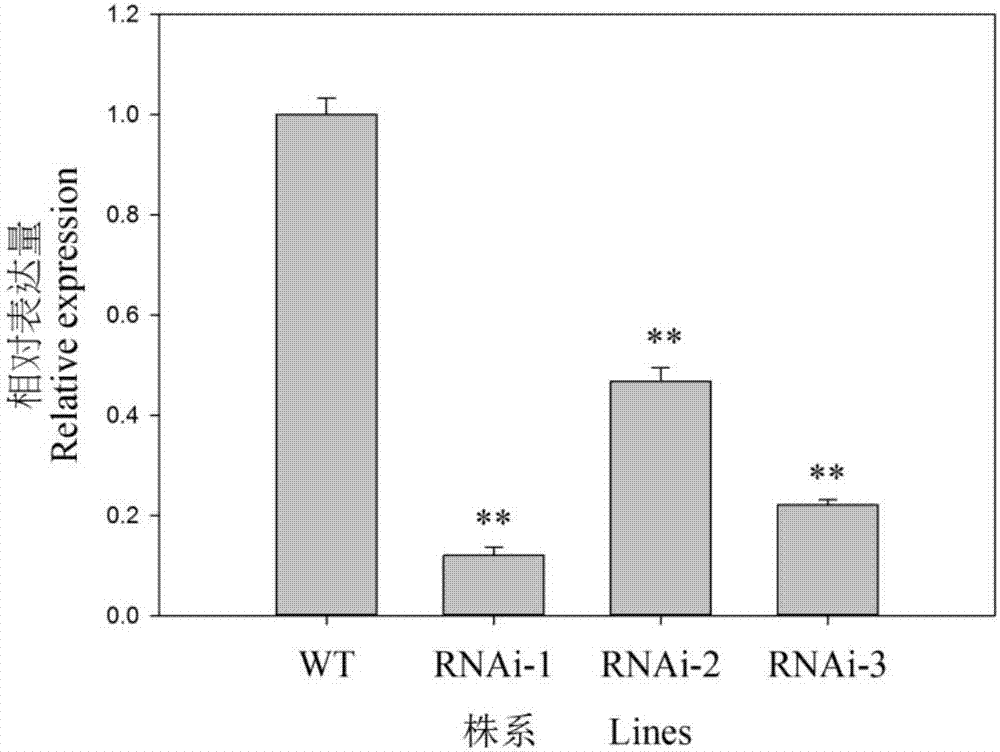
[0048] The OsbHLH116-RNAi expression vector was transformed into Agrobacterium EHA105, followed by Nishimura et al. (Nishimura et al., 2007) to infect the mature embryo callus of rice Xinfeng 2, and screened for hygromycin (Hyg) resistance to obtain regenerated seedlings. GUS staining was used to screen positive plants, and T 2 Generation of transgenic positive plants (OsbHLH116-RNAi-1, OsbHLH116-RNAi-2, OsbHLH116-RNAi-3 lines) were used for subsequent experiments.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com