Nucleic-acid extraction kit and application thereof
A kit and nucleic acid technology, applied in the field of molecular biology, can solve the problems such as the inability to extract a large amount of mortars, the inability to completely break the cell wall, and the inability to obtain a good wall removal effect, so as to be beneficial to removal, improve the lysis efficiency, and have a good effect. release effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] The preparation of embodiment 1 detection kit
[0055] (1) Preparation of reagents
[0056] Buffer A: 0.5-0.5M Tris-Cl buffer (pH5.0-8.0), 0.5-1.5M NaCl;
[0057] Enzyme solution B: 0.05-0.2M phosphate buffer (pH6.8-7.4), 0.25M MgCl2, 80-120mg / mL helicase;
[0058] Lysate C: 0.05-0.5M Tris-Cl buffer (pH5.0-8.0), 0.05-0.2M EDTA, 8-12% SDS, 8-12% Tween-40;
[0059] Suspension D: 5-40mg / ml RNAaseA
[0060] Remover E: 0.2-1M Al2(SO4)3, 80-350g / L CTAB, 0.5-1.5M NaCl;
[0061] Binding solution F: 0.1-0.5M Tris-Cl buffer solution (pH5.0-8.0), 2-8M guanidine isothiocyanate;
[0062] Wash solution G: 0.1-0.5M Tris-Cl buffer (pH5.0-8.0), 15-25% (volume ratio) absolute ethanol;
[0063] Eluent H: sterile deionized water;
[0064] (2) Consumables
[0065] Glass Beads, Spin Columns, and Collection Tubes
[0066] (3) Assembly of the kit
[0067] Assemble the primer composition, detection probe and reagents related to the kit to prepare the detection kit.
Embodiment 2
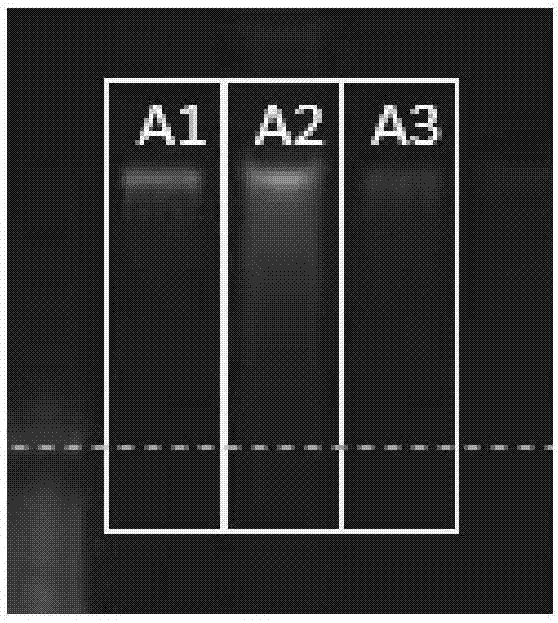
[0068] The extraction of embodiment 2 sample DNA (A1 / B1 / C1)
[0069] Select 3 stool samples: A, B, and C, divide them into 6 parts on average, and mark them as: A1-A6, B1-B6, C1-C6, and the three samples of A1, B1, and C1 are processed by the method of this embodiment. The DNA extraction method comprises the following steps:
[0070] (1) Take 800 μl of buffer A and 200 mg of glass beads into a 2 ml centrifuge tube;
[0071] (2) Add 200mg sample to the above 2ml centrifuge tube, vortex and mix for 30s;
[0072] (3) Add 100 μl enzyme solution B to the centrifuge tube in step (2), and incubate at 45-65°C for 20 minutes;
[0073] (4) Add 50 μl of lysate C to the sample, vortex for 5 minutes to mix the sample, centrifuge at 12,000 rpm (~13,400×g) for 60 s, transfer the supernatant (400 μl) to a new 2ml centrifuge tube;
[0074] (5) Add 300μl suspension D, vortex for 5-10s, place at room temperature for 5min, centrifuge at 12,000rpm (~13,400×g) for 60s, and precipitate the sample...
Embodiment 3
[0084] The extraction of embodiment 3 sample DNA (A1 / B1 / C1)
[0085] The three samples of A1, B1, and C1 are processed by the method of this embodiment, and the specific DNA extraction method includes the following steps:
[0086] (1) Take 800 μl of buffer A and 100 mg of glass beads into a 2 ml centrifuge tube;
[0087] (2) Add 200mg sample to the above 2ml centrifuge tube, vortex and mix for 30s;
[0088](3) Add 50 μl enzyme solution B to the centrifuge tube in step (2), and incubate at 40-50° C. for 120 minutes;
[0089] (4) Add 100 μl of Lysis Buffer C to the sample, vortex for 5 minutes to mix the sample, centrifuge at 12,000 rpm (~13,400×g) for 60 s, transfer the supernatant (400 μl) to a new 2ml centrifuge tube;
[0090] (5) Add 300μl suspension D, vortex for 5-10s, place at room temperature for 5min, centrifuge at 12,000rpm (~13,400×g) for 60s, and precipitate the sample particles;
[0091] (6) Transfer the supernatant to a new 2ml centrifuge tube, add 50μl remover ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com