Primer group and probe for detecting ostreid herpesvirus of infected scapharca subcrenata, and application of primer group and probe
A technology of oyster herpes virus and primer set, which is applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc. It can solve the problems of difficult design, high cost of RPA primers and probes, and achieve low requirements for equipment and rapid response , the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Design and specificity detection of embodiment 1 primer set and probe
[0024] According to the virus ORF 95 (GenBank accession no.AY509253.2) nucleic acid sequence design primer set and probe set, wherein the target gene sequence is as follows:
[0025]
[0026] Primer sets are:
[0027] Forward primer FP: catgtttacg tggaaatgtt ggattggcta 30
[0028] Reaction primer RP: atgtcaaata ggttgttggc agtgatggtc 30
[0029] The probe sequence is:
[0030] tacagcatcg cccgatgcac ttcgtgat(dT-FAM)ca(THF)tt(dT-BHQ1)atcggctcaatatata(C3spacer)
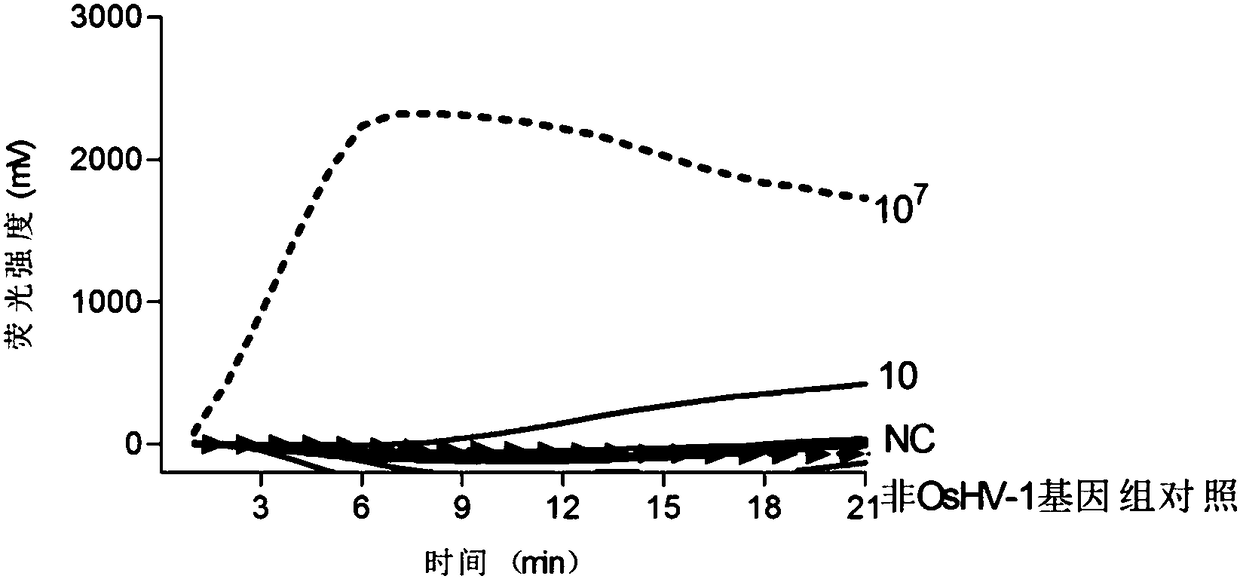
[0031] Specific detection: establish a positive control containing OsHV-1 plasmid, ddH 2 O negative control, shrimp virus (WSSV) genomic DNA control, abalone herpes virus (AbHV) control, grouper viral neuronecrosis virus (RGNNV) control, Harvey Vibrio genomic DNA control, Dongfeng snail genomic DNA control, The water body DNA control and the healthy prawn genome DNA control were detected using these genome DNA as templates.
[0032] Gr...
Embodiment 2
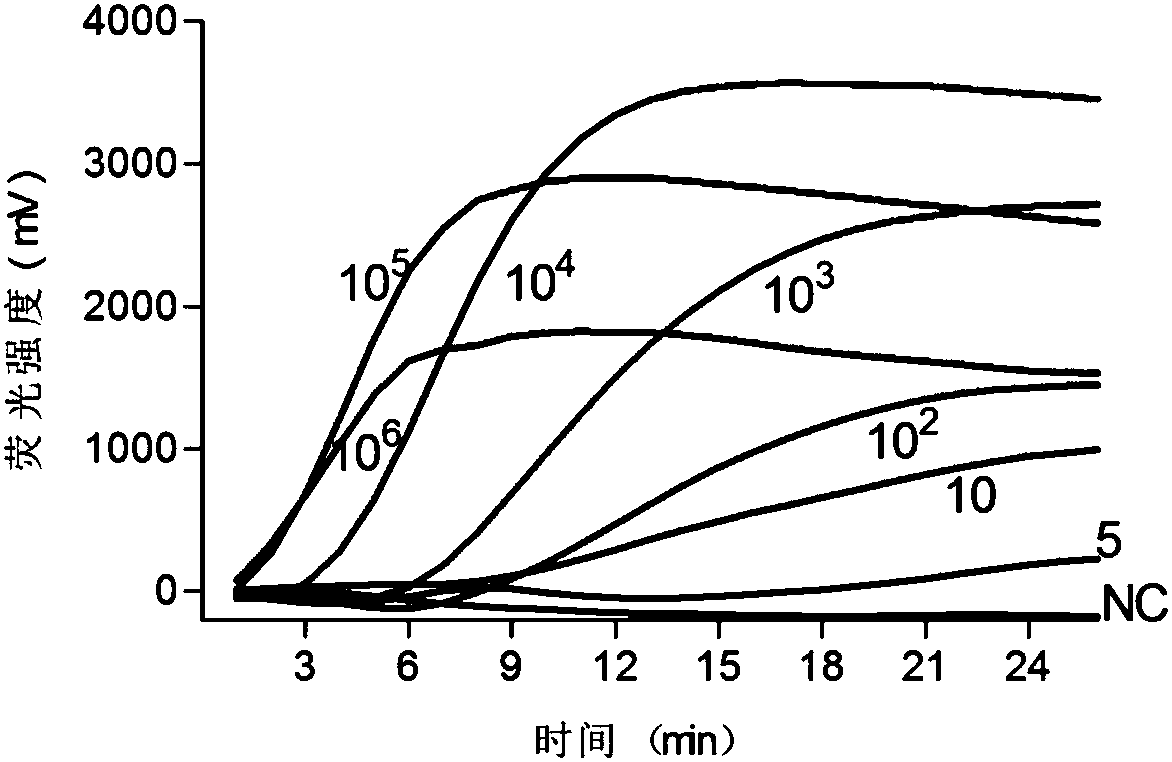
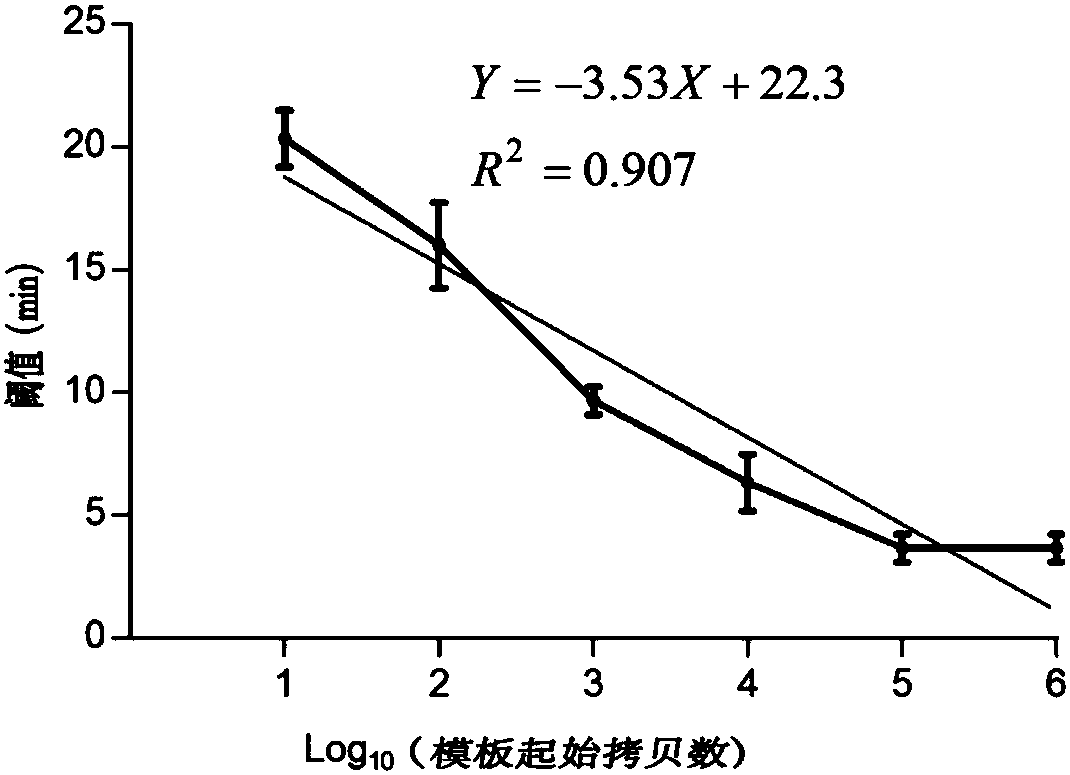
[0044] Example 2 Sensitivity Verification
[0045] RPA products were used to construct recombinant plasmids. The plasmid is formed by connecting the target gene fragment and the PMD18-T vector.
[0046] The vector sequence has been made public: http: / / wenku.baidu.com / view / 385564bdf121dd36a32d8271.html.
[0047] Detection by absolute quantification method: the constructed plasmid standard was treated with ddH 2 O was diluted 10 times to construct different concentration gradients of 10 6 、10 5 、10 4 、10 3 、10 2 , 10 and 5 to make a standard curve, use ddH 2O was used as a negative control, and each concentration was repeated three times. The RPA amplification reaction was performed using a real-time fluorescent quantitative PCR instrument (Eppendorf).
[0048] reaction system:
[0049] Rehydration buffer (Twistdx kit, TAEXO02KIT): 14.7μL
[0050] RPA FP (10 μM): 1.1 μL
[0051] RPA RP (10 μM): 1.1 μL
[0052] Probe (10μM): 0.3μL
[0053] f 2 O: 4.6 μL
[0054] D...
Embodiment 3
[0060] Embodiment 3 Feasibility verification
[0061] In order to further confirm the reliability of the qRPA detection system of the present invention, we randomly selected 50 cockles, and simultaneously performed qRPA and qPCR detection of the OsHV-1 content in their bodies. The clam DNA was extracted using a mollusk animal extraction kit (Magen, Guangdong, China), and the specific operation was performed according to the instructions.
[0062] The qRPA detection method, primer set and probe are the same as in Example 2.
[0063] The qPCR detection method used the reported SYBR Green detection method (Pepin, Riou et al. (2008). "Rapid and sensitive detection of ostreid herpesvirus 1 in oystersamples by real-time PCR". Journal of Virological Methods). The construction method of qPCR bracket is similar to the construction method of qRPA bracket in Example 2.
[0064] The primer sequences used are:
[0065] C9: GAGGGAAATTTGCGAGAGAA
[0066] C10: ATCACCGGCAGACGTAGG
[0067]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com