Phospholipase and application thereof
A technology of phospholipids and polynucleotides, applied in the field of phospholipases, can solve problems such as ineffectiveness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0119] Embodiment 1: phospholipase error-prone PCR of the present invention
[0120] EPCR system (50ul):
[0121]
[0122] Using SEQ ID NO: 1 as a template, using the sequence shown in SEQ ID NO: 3 as an upstream primer and the sequence shown in SEQ ID NO: 4 as a downstream primer to perform error-prone PCR.
Embodiment 2
[0123] Embodiment 2: Construction of phospholipase mutant library of the present invention
[0124] Use 1% agarose (purchased from AMRECSO) gel electrophoresis to separate and purify the error-prone PCR product target band, and use the E.Z.N.ATM gel recovery kit from OmegaBio-Tek, USA to recover the target band. The products recovered from the gel were digested with two restriction endonucleases AvrII and NotI-HF, and then the digested products were recovered with the E.Z.N.ATM product recovery kit from OmegaBio-Tek Company. Simultaneously carry out enzyme digestion of pPIC9K vector and recovery of enzyme digestion products.
[0125] Take an appropriate amount of the above two recovered enzyme digestion products, mix them, add a certain amount of ligase and ligase buffer, and place them in a water bath at 22°C for about 2 hours to react.
[0126] Take out one tube containing 100 μl of DH5α competent cells, after the ice bath melts, add 20 μl of the ligation product accordingl...
Embodiment 3
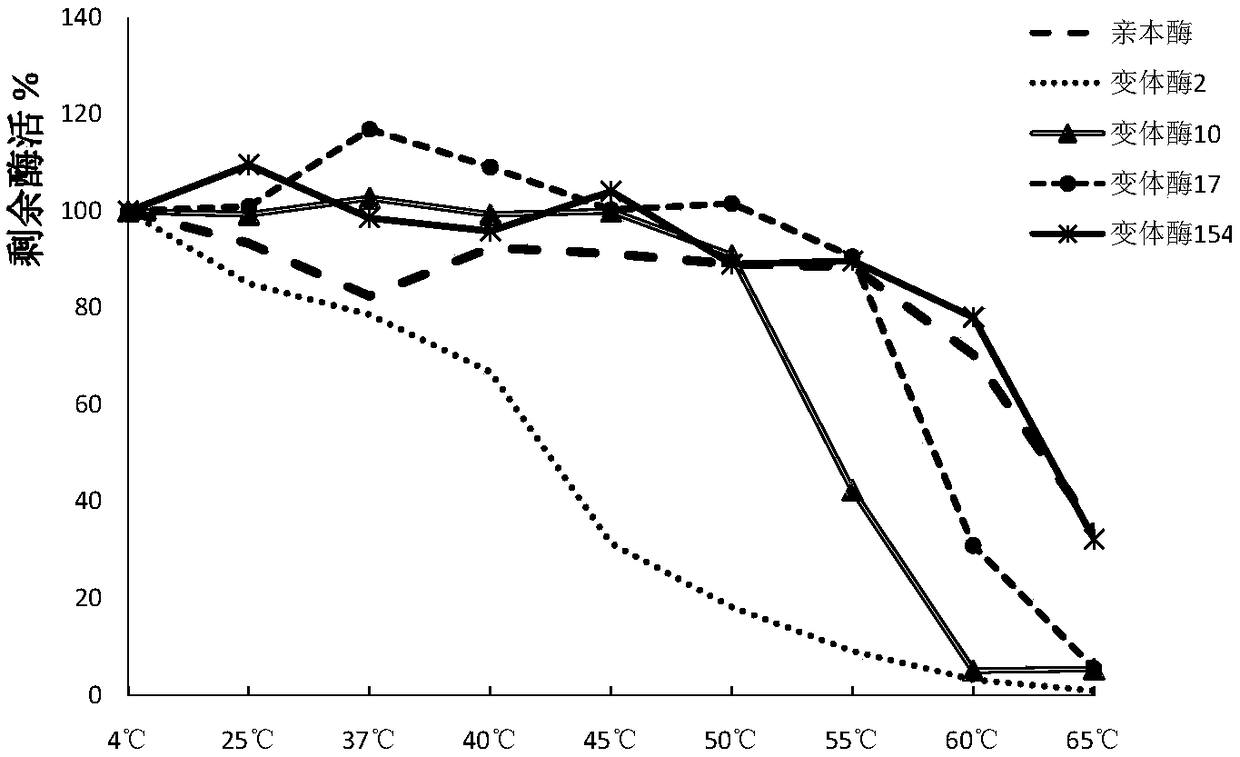
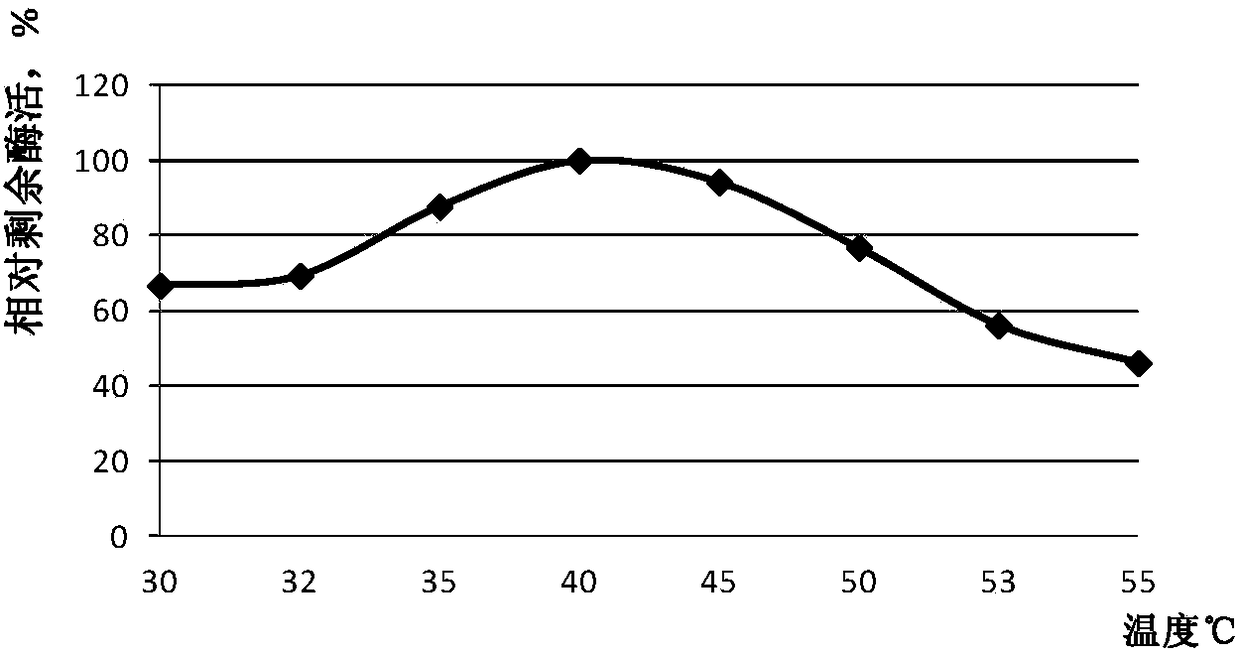
[0132] Example 3: Plate Screening of Phospholipase Mutant Library
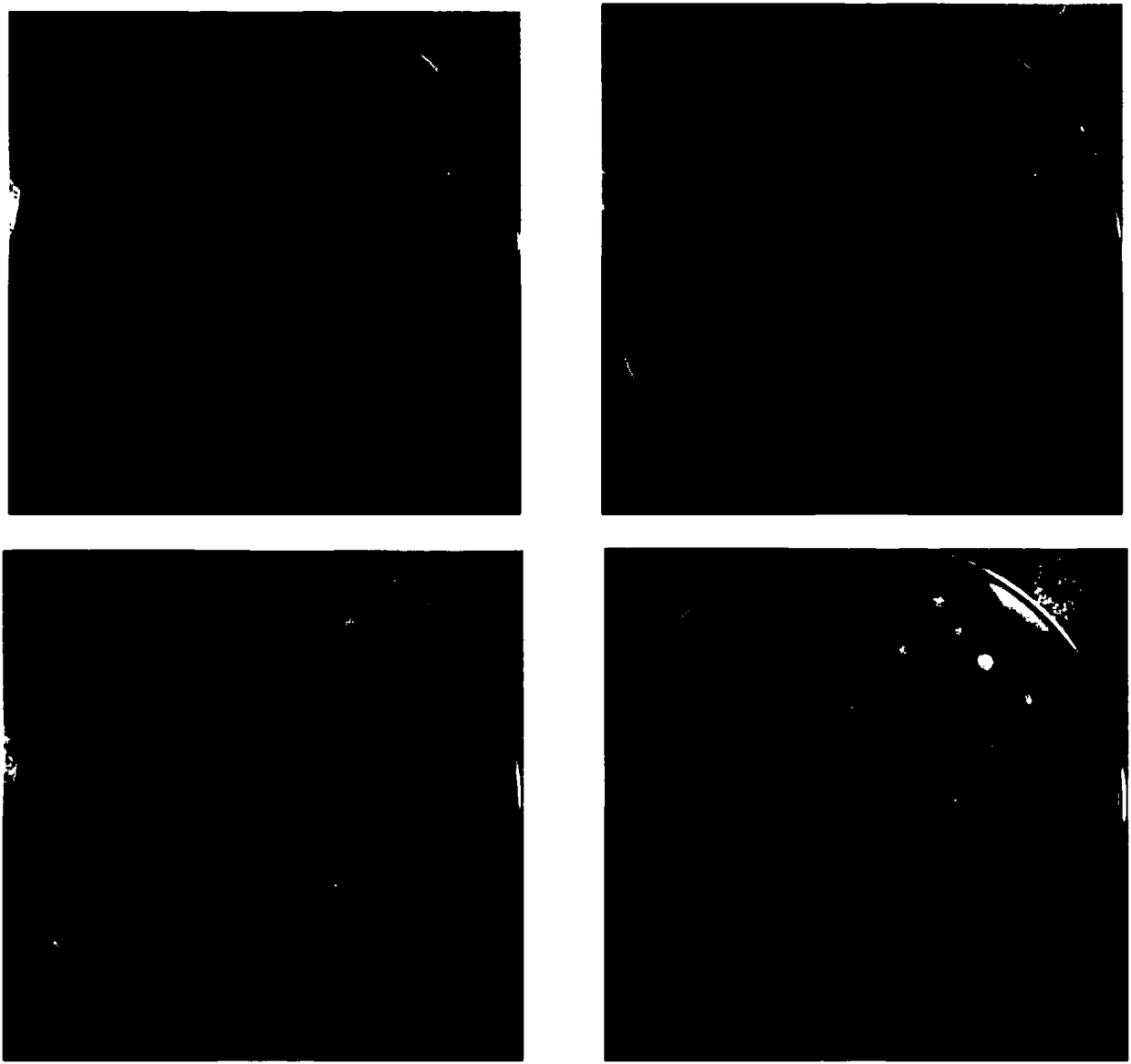
[0133] Plate all the bacteria of ECSPLB-pPIC9K-GS115 mutant onto BMMY plate (1% yeast extract, 2% peptone, 100mM potassium phosphate pH6.0, 1.34% YNB 4×10-5% biotin, 0.5% methanol, 1 % soybean lecithin, 1.5% agar), cultured at 30°C for 3 days, the results were as follows figure 1 shown. figure 1 Among the four plates, one bacterium in the middle of each plate is the phospholipase parent, and the other bacteria are phospholipase variants. Pick forward phospholipase variants with larger clear and cloudy circles than the parental circle. figure 1 In the middle, the black square marked in the upper left picture is the bacteria containing phospholipase variant 154, the black square marked in the upper right picture is the bacteria containing phospholipase variant 2, and the black square marked in the lower left picture is the bacteria containing phospholipase variant 10 Bacteria containing phospholipase variant ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com