A Hong Kong oyster lysm protein with antibacterial activity and its coding gene and application
A Hong Kong oyster and protein technology, applied in Hong Kong oyster LysM protein and its coding gene and application field, can solve the problems of breeding oyster death, economic loss, and threats to the sustainable development of existing industries, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
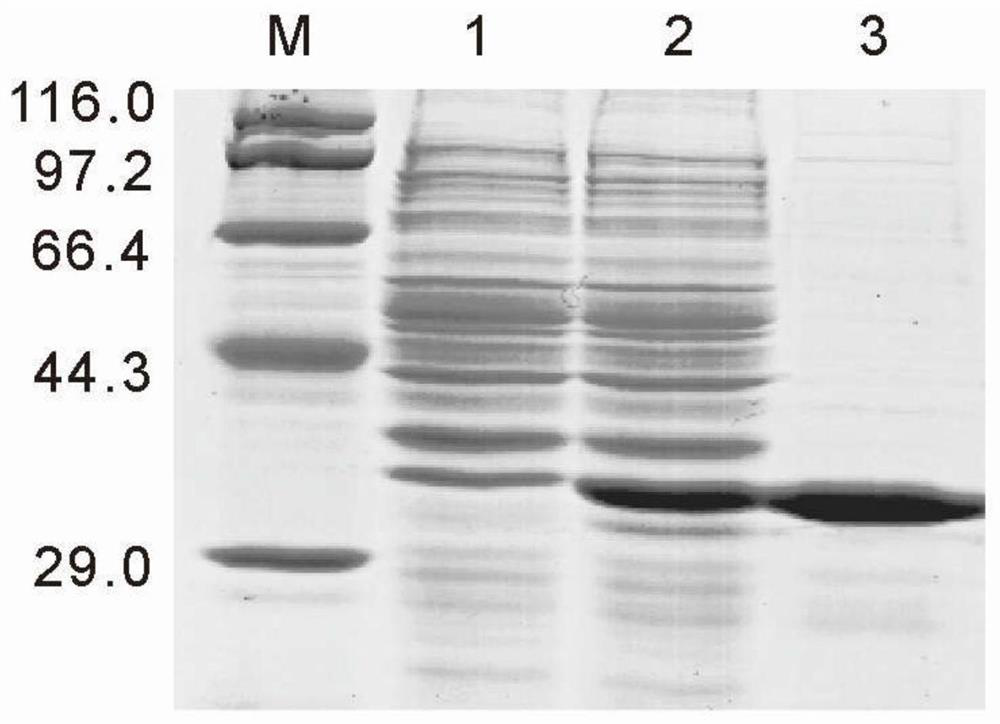
[0020] In Vitro Recombinant Expression and Purification of LysM Protein from Hong Kong Oyster
[0021] The amino acid sequence of the Hong Kong oyster LysM protein is shown in SEQ ID NO.1:
[0022] MSSRKSRQDANNKPYSYQQLGAEVQNSKKSRVYVFGNADVQEDEVVEFEMSDVRSR KGGPKPTQKDEDEQLYYERELTEGDTLRSLSLQYGCPVAEIKRINNMIQDQDFYAYRKIKVP IKKYSFLTETLKTETKSDKPQFNGITVVDETDTENTETDSESVCNMSDPETQRLMIKKSLSIR SQTGLQSKEARRFLRSMDKDLVKICKSAKMRHESLDEVVSLLTNRSIQPIPPPRRKFFGHDC GLTWCSMIGVIVVLAILIPLGILSYLWFTGHFSTHSSAGQNNG
[0023] Length: 286 amino acids
[0024] Type: protein
[0025] Chain type: single chain
[0026] Topology: Linear
[0027] Features: The molecular weight is 32.7kDa, the isoelectric point is 8.16, and its coding sequence has a conserved LysM domain
[0028] Source: Hong Kong Oyster (Crassostrea hongkongensis)
[0029] 1. Construction of prokaryotic expression vector of LysM gene of Hong Kong oyster: (1) design a pair of primers F1 covering ChLysM (LysM gene of Hong Kong oyster) ORF (its nuc...
Embodiment 2
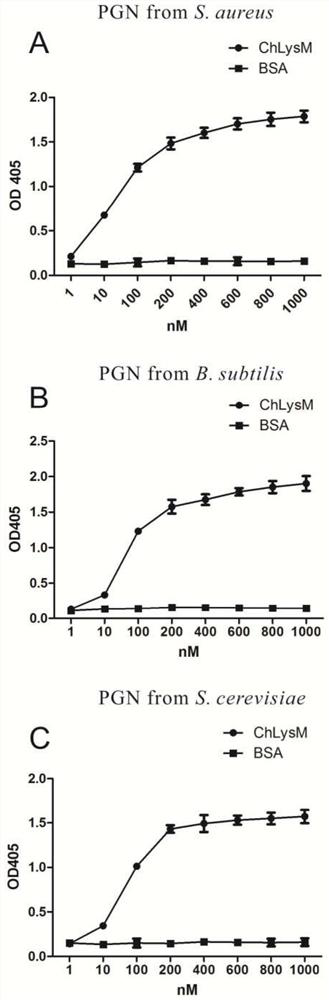
[0033] Hong Kong oyster LysM protein recognizes peptidoglycan (PGN) from different sources
[0034] (1) Dissolve PGN from three sources of staphylococcus aureus (Sigma, 77140), Bacillus subtilis (Sigma, 69554), and Saccharomyces cerevisiae (Sigma, 72789) in double distilled water (40mg / ml); (2) respectively Pipette 50 μl into a 96-well plate (Costar), and place at room temperature until evaporated to dryness; (3) fix PGN at 60°C for half an hour; (4) add 200μl Tris Buffer containing BSA, 37°C, 2h; (5) discard Supernatant, Tris Bufferwash 4 times, 2min each time; (6) Add purified ChLysM recombinant protein at different concentrations, and incubate at room temperature for 3h; (7) Discard the supernatant, Tris Buffer wash 4 times, 2min each time; (8) Add mouse anti-GST antibody (Abmat, 1:5000), incubate for 2 h; (9) discard the supernatant, Tris Buffer wash 4 times, 2 min each time; (10) add HRP-coupled goat anti-mouse IgG (Abmat, 1 :3000), incubated for 2h; (11) adding HRP subs...
Embodiment 3
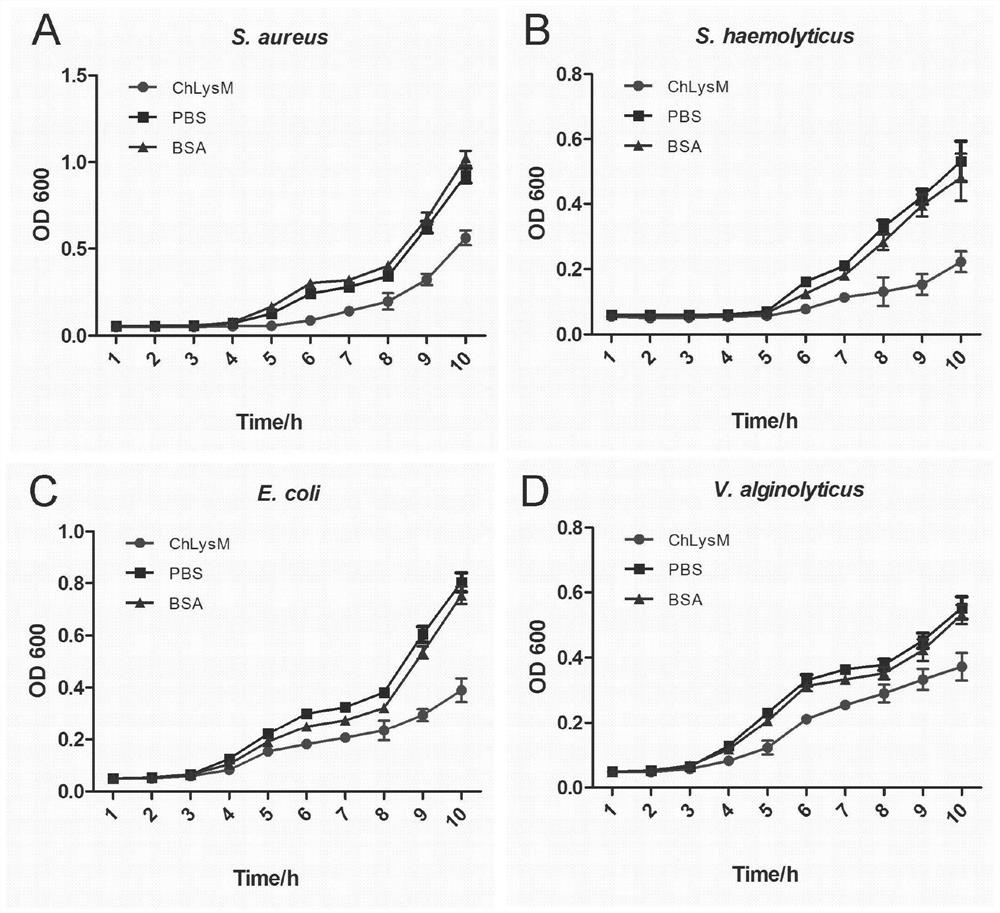
[0037] Antibacterial activity of LysM protein from Hong Kong oyster
[0038](1) Vibrio alginolyticus (Vibrio alginolyticus), staphylococcus hemolyticus (staphylococcus aureus), staphylococcus aureus (staphylococcus aureus) and Escherichia coli (Escherichia Coli) four kinds of bacteria use LB culture medium at respective suitable temperature, 220rpm, cultivate to During the logarithmic growth phase; (2) Dilute the bacteria with Tris-HCl (50mmol L-1, pH=8.0) so that the number of colonies per milliliter of the bacteria solution is about 1×10 4 CFU; (3) 50 μL of Hong Kong oyster LysM protein (100 ng / μL) was incubated with 50 μL of the above bacterial dilution at 30° C. for 3 h. (4) Take 20 μL of the above mixture in a flat-bottomed 96-well plate (Costar), add 200 μL LB liquid medium to each well, continue culturing, and measure and record the OD600 value of each well every 1 h. (5) Set up BSA protein control group and PBS blank control group. Three repetitions are set for each ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com