A quantum dot-magnetic bead miRNA sensor based on dsn enzyme and its preparation method and detection method
A quantum dot and sensor technology, applied in the field of nano-biomedicine, can solve the problem that single-stranded DNA and RNA have no effect, and achieve the effect of wide linear range, high operation sensitivity, and high detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Add 22.5 μL of SA-QD (Streptavidin QD) with a concentration of 1 μM, 22.5 μL of Biotin-DNA with a concentration of 10 μM, 200 μL of blocking solution, and 600 μL of Tris-HCl buffer solution into a 1.5 mL low adsorption centrifuge tube and shake at a speed of 6000 rpm / min for 1 hour , construct the QD-DNA probe, and then ultrafilter at 4000rpm for 5min to remove the DNA not connected with QD, and repeat the ultrafiltration process 3 times. After the last ultrafiltration until 100 μL of the QD-DNA probe solution remained, 75 μL of Tris-HCl buffer and 50 μL of blocking solution were added to obtain a high-concentration QD-DNA probe. Divide the constructed high-concentration QD-DNA probe into 10 equal parts: take 22.5 μL QD-DNA probe for each part, add 135 μL Tris-HCl buffer solution and 67.5 μL blocking solution.
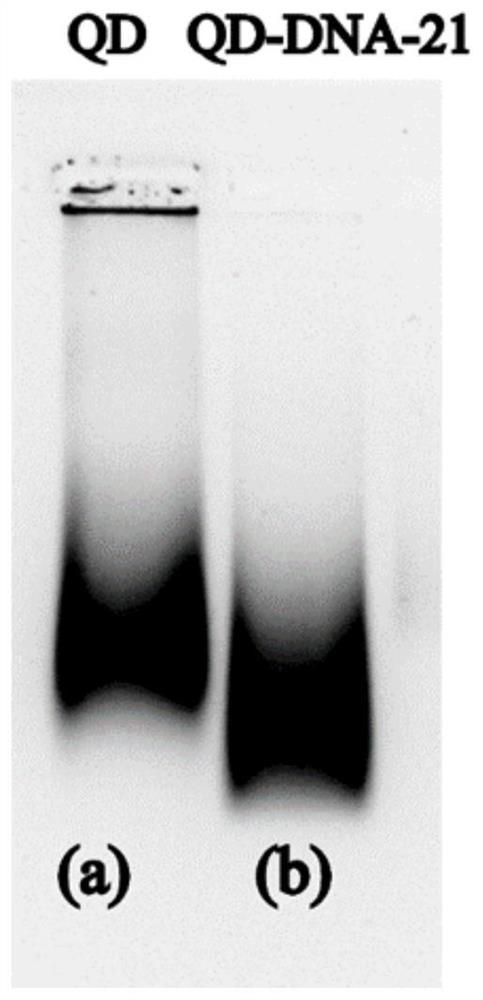
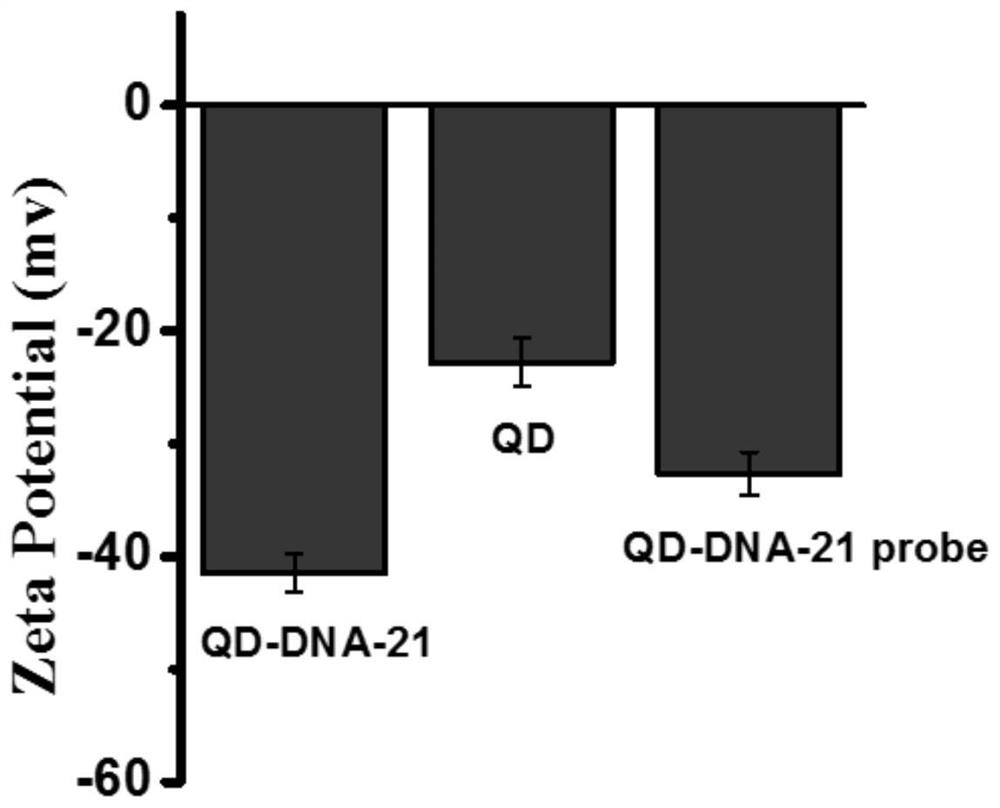
[0049] The surface of quantum dots is negatively charged, and the surface of DNA is negatively charged. After QD is connected to DNA, the negative charge densit...
Embodiment 2
[0051] Before the experiment, wash the MB with ligation-washing buffer (B&W Buffer) 3 to 4 times to remove substances such as sodium azide in the mother liquor, take 45 μL of MB with a concentration of 10 mg / mL, 45 μL of Biotin-DNA with a concentration of 10 μM, and 600 μL Tris-HCl buffer solution was added to a low-adsorption centrifuge tube and shaken at a speed of 6000 rpm / min for 2 hours to construct the MB-DNA probe. Because the particle size of MB is relatively large (particle size is 1 μm), sedimentation is easy to occur, so the solution in the centrifuge tube was mixed every 30 minutes, so that MB and DNA can be fully connected. The constructed MB-DNA probe was magnetically separated and washed three times, and 225 μL of Tris-HCl buffer was added to obtain a high-concentration MB-DNA probe. Divide 10 equal parts of the MB-DNA probe solution: take 22.5 μL of high-concentration MB-DNA probe solution for each part, and then add 227.5 μL of hybridization buffer to make the...
Embodiment 3
[0054] Take 12 μL of 1 μM SA-QD (Streptavidin QD), 30 μL of Biotin-DNA with a concentration of 10 mM, 200 μL of blocking solution, and 600 μL of Tris-HCl buffer, and add them to a 1.5 mL low-adsorption centrifuge tube and shake at 6000 rpm / min for 2 hours , to construct QD-DNA bioprobes. Transfer the ligation product to an ultrafiltration tube and ultrafilter at a speed of 4000rpm for 5 minutes to remove as much as possible the DNA that is not connected to the QD in the supernatant. Ultrafilter three times in a row, take out the ultrafiltration tube and mix the supernatant with a pipette Prevent QDs from sticking to the filter membrane. Take out the supernatant and dilute the solution to 550 μL with ultrapure water, and prepare a QD solution of the same concentration with ultrapure water.
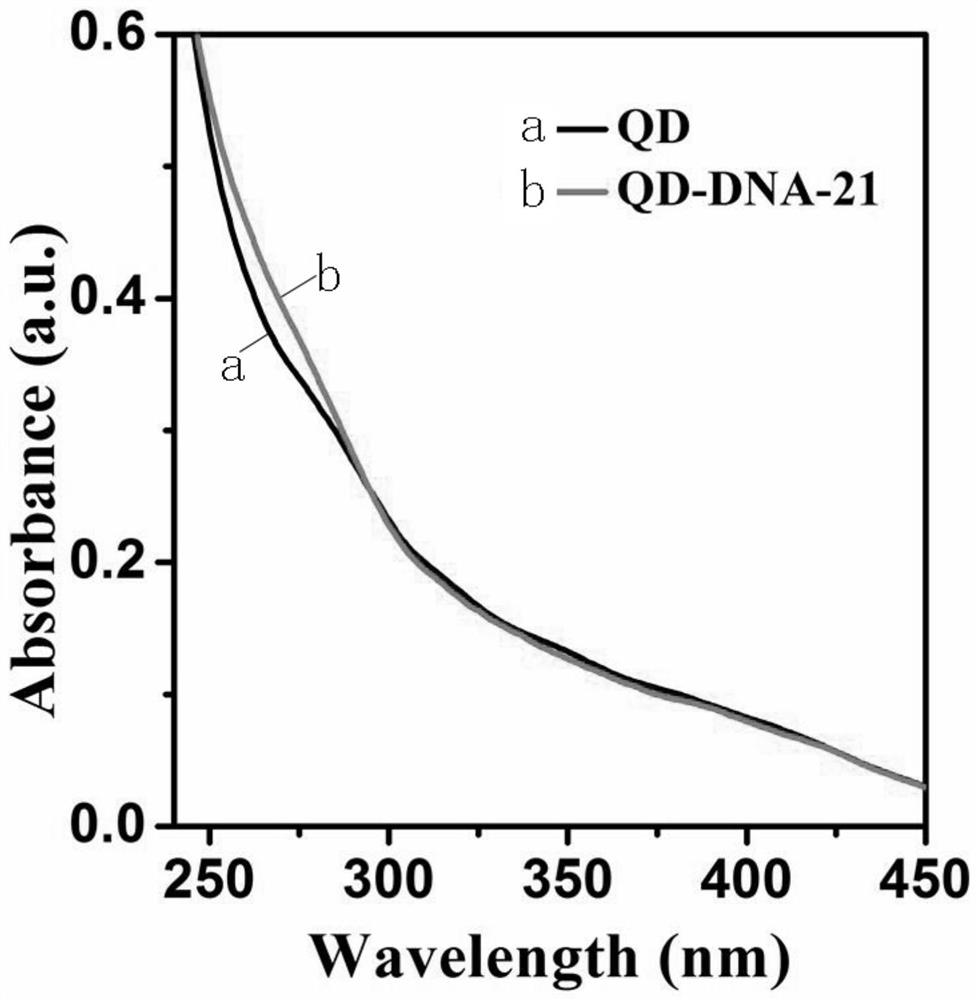
[0055] DNA and QD are coupled together by streptavidin and biotin, and the characteristic absorption peak of DNA can be observed by UV characterization of QD-DNA probe. From figure 2 It...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com