Amino lyase mutant protein and coding gene and application thereof
An amino acid and protein technology, applied in the directions of lyase, application, genetic engineering, etc., can solve the problems of complex chemical synthesis process, low overall yield and high production cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
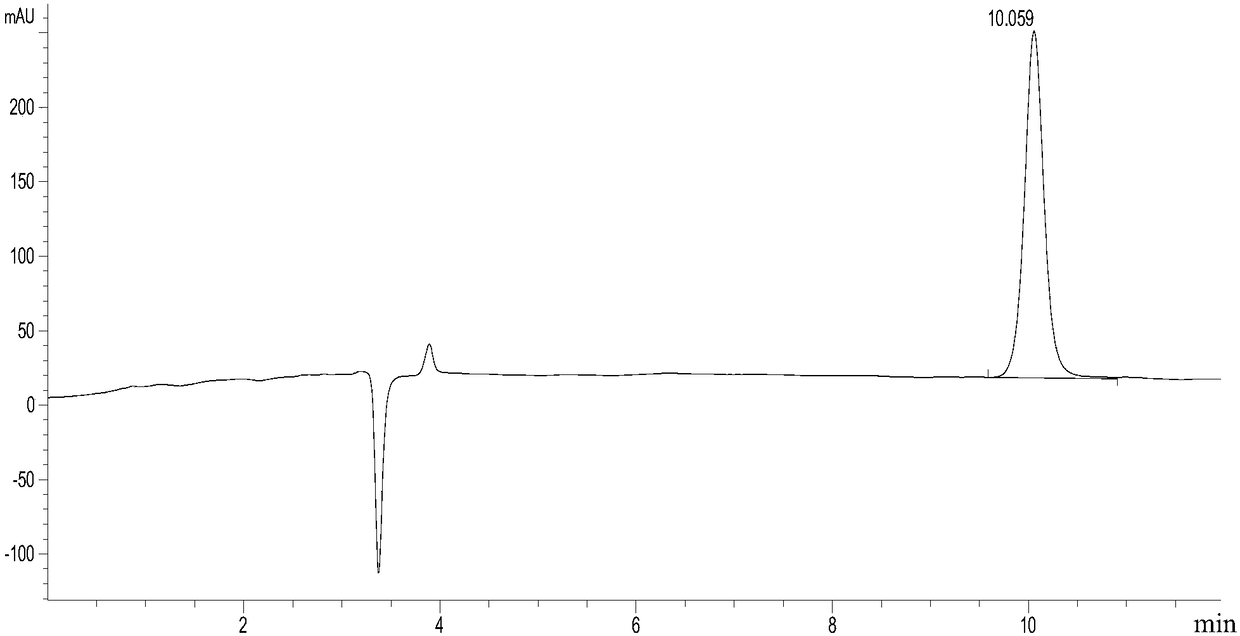
[0133] Example 1. Screening and determination of mutation sites
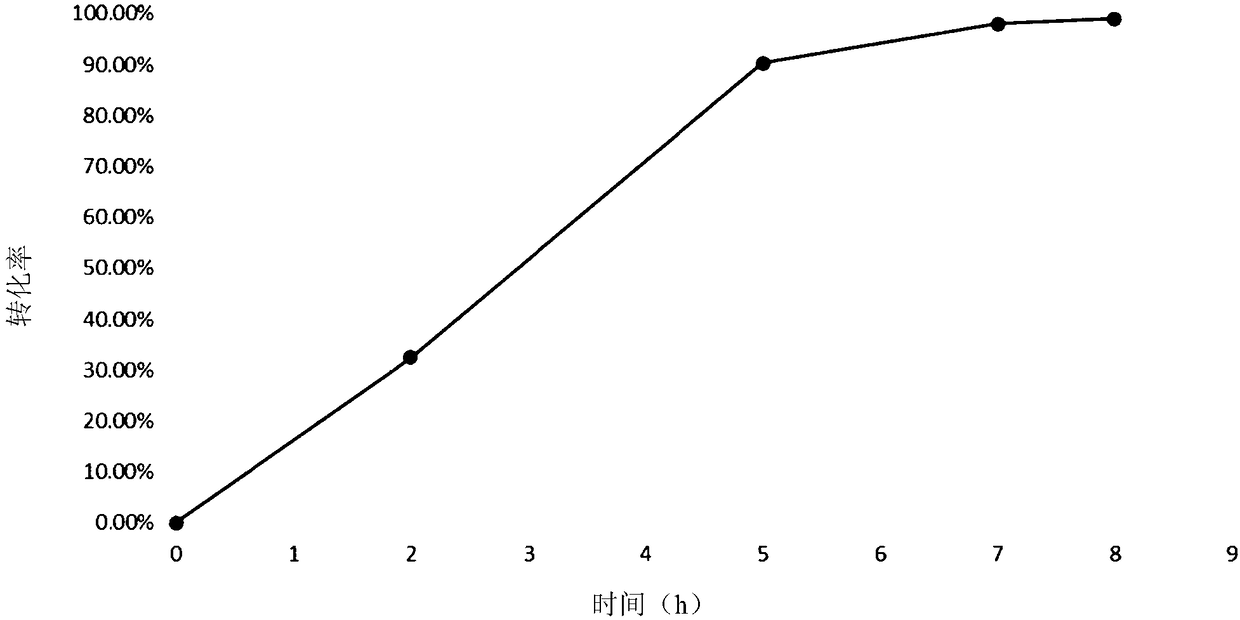
[0134] Sequence analysis, mutation and functional verification of L-aspartate deaminase protein (aspB protein) derived from Bacillus subtilis found 37 amino acid sites. Four important amino acid sites were screened. These 4 amino acid sites are mutated in different forms, and the obtained mutant proteins all have higher aminolyase activity, can catalyze the ammonia addition reaction with trans-2-butenoic acid as a substrate simultaneously, and can be used for the production of ( R) 1-propyl(2-amino)carboxylic acid in high yield and meeting the requirement of 100% stereoselectivity.
[0135] The aspB protein is shown in SEQ ID NO: 2 in the sequence listing, and the encoding gene (aspB gene) of the aspB protein is shown in SEQ ID NO: 1 in the sequence listing.
[0136] The four amino acid sites and their mutant forms are shown in Table 1.
[0137] Table 14 amino acid sites and their mutated forms
[0138]
Embodiment 2
[0139] Embodiment 2, the preparation of recombinant bacteria
[0140] The wild-type aspB protein is shown in SEQ ID NO: 2 of the Sequence Listing, and the encoding gene (aspB gene) of the wild-type aspB protein is shown in SEQ ID NO: 1 of the Sequence Listing.
[0141] 1. Construction of wild-type recombinant expression vector
[0142] The double-stranded DNA molecule shown in sequence 1 was inserted between the EcoR I and Not I restriction sites of the pET21a vector, and the recombinant expression vector pET21a-aspB was obtained (sequencing verification was correct). The DNA molecule shown in SEQ ID NO: 1 encodes the protein shown in SEQ ID NO: 2.
[0143] 2. Construction of mutant recombinant expression vector
[0144] 1. Insert the double-stranded DNA molecule 1 between the EcoR I and Not I restriction sites of the pET21a vector to obtain the recombinant expression vector pET21a-1 (sequencing verification is correct). Double-stranded DNA molecule 1 is obtained by point m...
Embodiment 3
[0181] Example 3. Determination of enzyme activity of aminotransferase mutant protein
[0182] The wild-type recombinant bacteria and mutant recombinant bacteria 1-34 obtained in Example 2 were used to carry out the following experiments respectively:
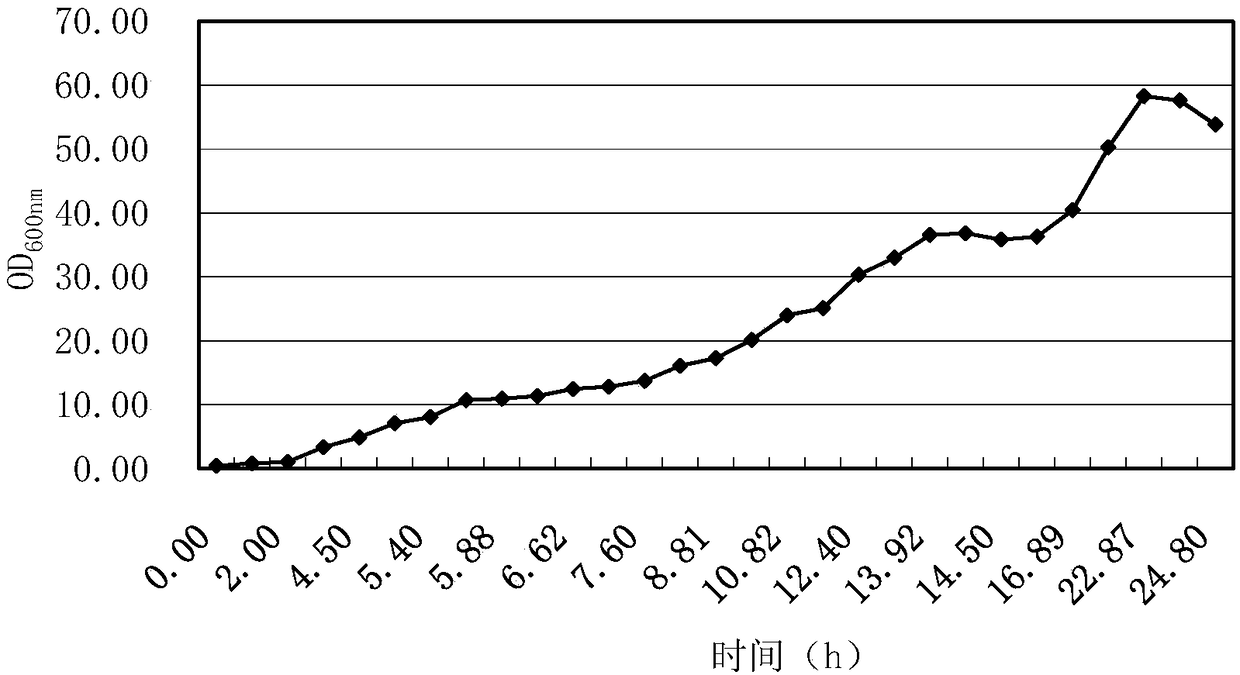
[0183] 1. Inoculate the bacteria to be tested in 5ml LB liquid medium (ampicillin resistance: 100μg / ml), shake at 37°C and 200rpm to the OD of the bacteria liquid 600nm = 1-2, add 30 ppm IPTG, continue to 30 ℃, 200 rpm shaking culture to 24 h.
[0184] 2. After completing step 1, centrifuge at 12,000 rpm to collect the bacteria.
[0185] 3. Use 50 mM Tris (pH 7.5), 2 mM MgCl for the cells collected in step 2 2 After 1ml of resuspended, ultrasonic (power of 25W) was broken for 30s to obtain the broken liquid of bacterial cells.
[0186] 4. The bacterial cell fragmentation solution obtained in step 3 was heated at 60° C. for 30 min, and then centrifuged at 12,000 rpm to collect the supernatant, which was the protein solution....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com