Anchoring polypeptide taking basic amino acid as anchoring end and application of anchoring polypeptide
An acidic amino acid and amino acid technology, applied in peptides, fluorescence/phosphorescence, instruments, etc., can solve the problems of low cysteine loading efficiency and time-consuming, and achieve excellent protection performance, less dosage, and enhanced stability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] (1) Design of anchoring polypeptide
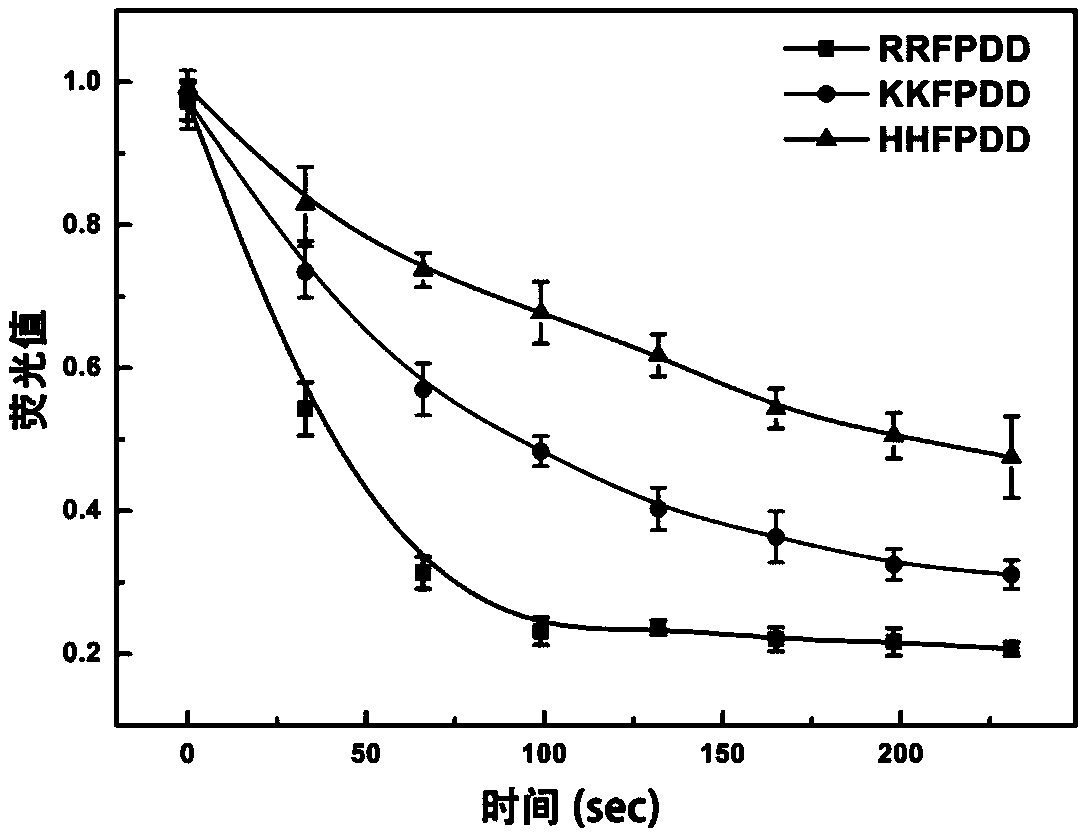
[0042] Such as figure 1 As shown, three kinds of polypeptide sequences (Sequence 1, Sequence 2, Sequence 3) with basic amino acids (arginine, lysine, histidine) as anchor ends were designed, and fluorescent groups were modified on their protected ends 6-carboxyfluorescein, the three polypeptide sequences and their modified 6-carboxyfluorescein are expressed as follows:
[0043] serial number
Polypeptide sequences anchored by basic amino acids
Polypeptide modified with 6-carboxyfluorescein
sequence 1
RRFPDD
RRFPDD-FAM
sequence 2
KKFPDD
KKFPDD-FAM
sequence 3
HHFPDD
HHFPDD-FAM
[0044] (2) Preparation of gold nanoparticles (AuNPs)
[0045] Add 100mL concentration of 0.01% (w / w) chloroauric acid aqueous solution into the flask and heat to boiling, then add 1.5mL trisodium citrate aqueous solution (33.8mM) into it rapidly under vigorous stirring, when the color of the s...
Embodiment 2
[0048] Example 2: Optimization of anchoring polypeptides
[0049] (1) Selection of anchor terminal amino acids
[0050] Fluorescence signals at different time points of the three AuNPs-polypeptide-FAM dispersions prepared in Example 1 were obtained by a fluorometer, and fluorescence kinetic curves were prepared. Due to the fluorescence quenching effect of gold nanoparticles, the most efficient basic amino acids can be screened out according to the fluorescence kinetic changes of the three AuNPs-polypeptide-FAM dispersions. The result is as figure 1 As shown, arginine (R) among the three basic amino acids can bring faster and stronger fluorescence signal changes, that is, bring higher polypeptide loading efficiency. Therefore, we choose arginine as the anchor end of the polypeptide to promote the modification efficiency of the polypeptide.
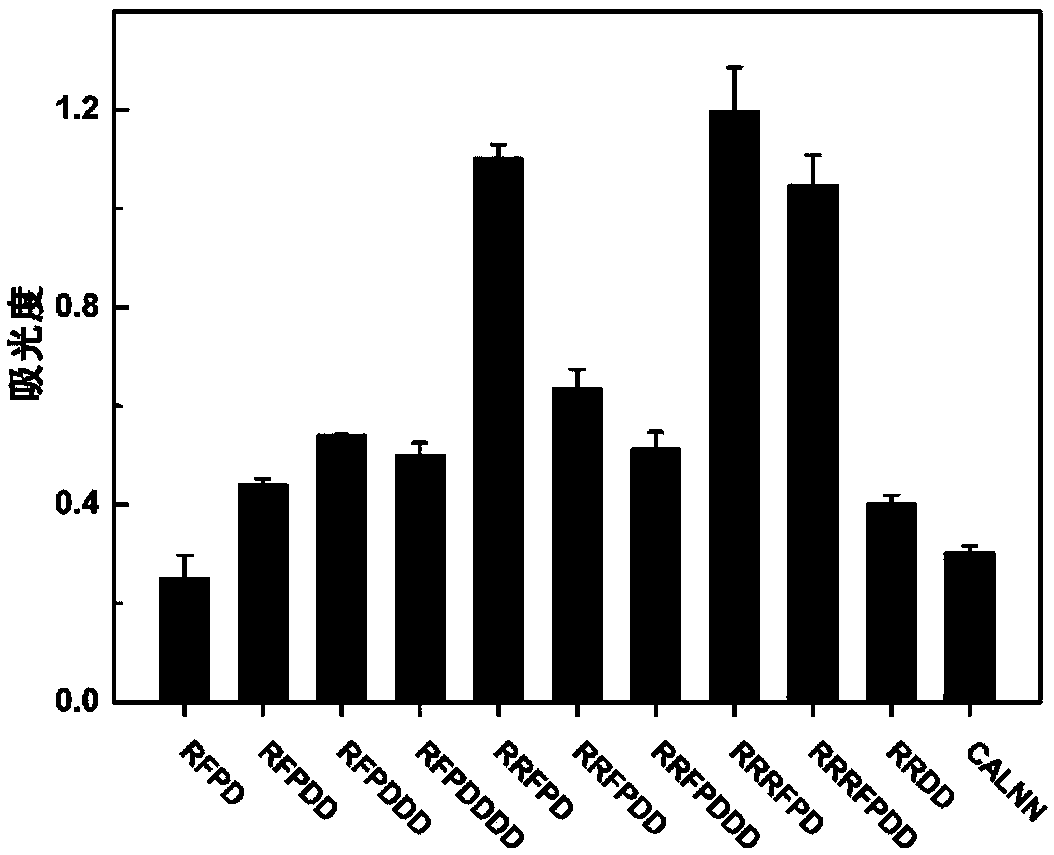
[0051] (2) Selection of the number of amino acids
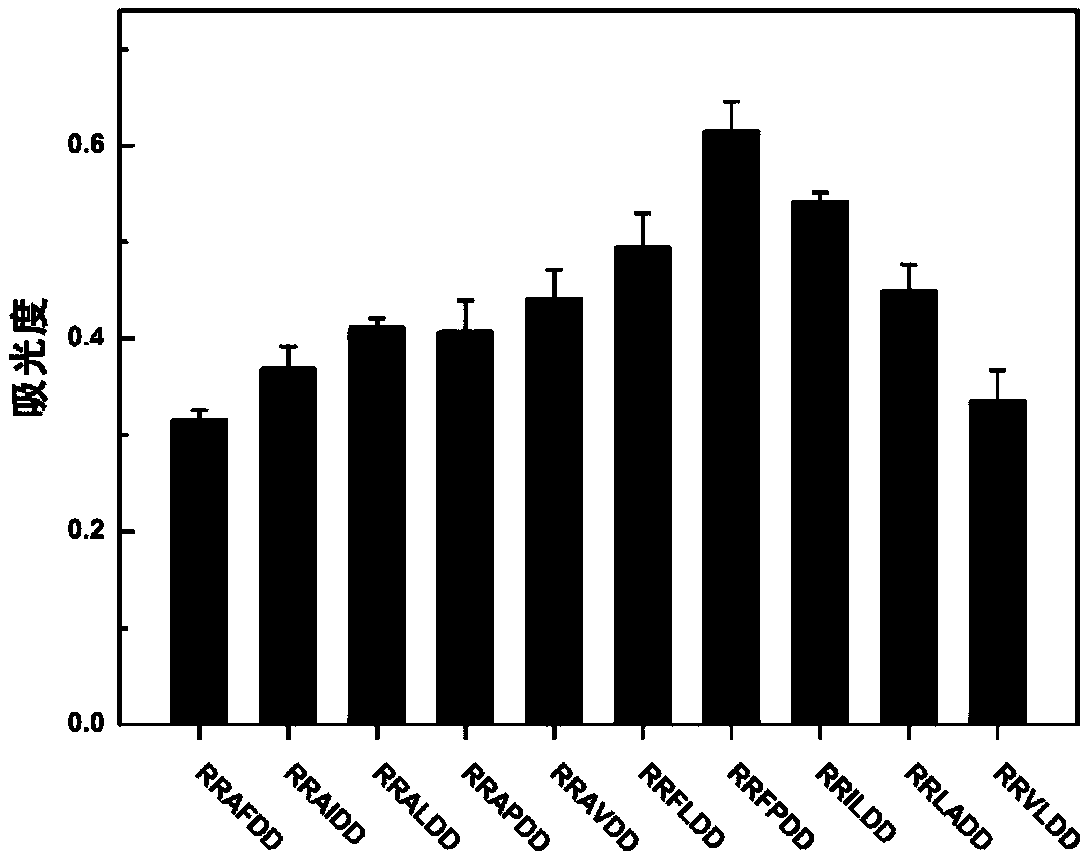
[0052] In addition, eight kinds of polypeptide sequences were designed and compared ...
Embodiment 3
[0067] (1) The anchor polypeptide RRFPDD connected with the target DNA, namely DNA-RRFPDD, was dissolved in NaH 2 PO 4 -Na 2 HPO 4 In the buffer solution (pH=7.4), the concentration of DNA-RRFPDD was 50 μM; then it was added to the AuNPs-phosphate dispersion, incubated at 37°C for 100 seconds and then centrifuged three times to remove excess DNA-RRFPDD, and the anchored polypeptide mediated Guided DNA-gold nanoparticles.
[0068] (2) Investigation of biological activity:
[0069] The hydrolysis performance of the polypeptide: select a target polypeptide with a fluorescent dye (sequence 26: RCFRGGDD, and load it on the gold nanoparticles by anchoring the polypeptide RRFPDD. After 100 seconds of loading, centrifuge and resuspend, add 100nM pancreatic Protease, detect fluorescence after incubation for 30 minutes. Through the change of fluorescence signal brought by enzymatic hydrolysis, verify whether its biological activity exists. Then by adjusting the modification ratio of...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com