Sodium/hydrogen antiporter gene sdmlt of Bacillus halophilus and its identification
A technology of antiporter and bacillus, applied in the field of Bacillus halophilus sodium/hydrogen antiporter gene sdmlT and identification, can solve the problems of unknown specific function, no guiding direction, lack of guiding significance, etc., and achieve enrichment of microbial salt tolerance Alkali gene resources, effect of improving salt-alkali tolerance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
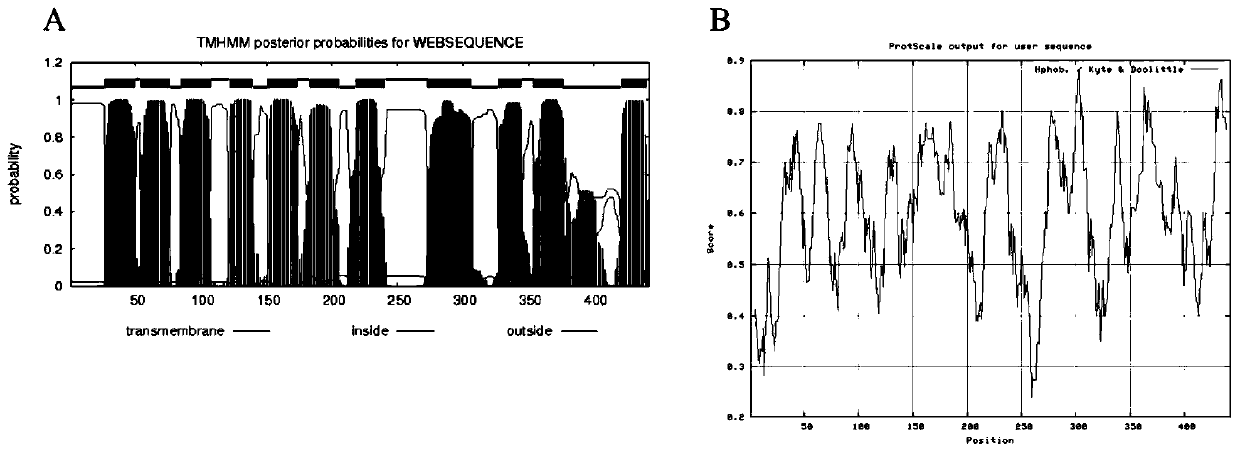
[0080] Example 1. Online biological software analyzes the transmembrane region and hydrophobicity of SdmlT protein, such as figure 1 Shown
[0081] The present invention uses online prediction websites http: / / www.cbs.dtu.dk / services / TMHMM-2.0 / and http: / / web.expasy.org / protscale / to perform bioinformatics analysis on the protein sequence encoded by the sdmlT gene, It was found that the protein SdmlT encoded by this gene has 11 transmembrane regions (as shown in Table 1), and the protein exhibits high hydrophobicity.
[0082] Table 1 Putative transmembrane region of SdmlT protein
[0083]
[0084] figure 2 It is the comparison of the amino acid sequence of SdmlT and the homologue provided in the embodiment of the present invention;
[0085] Ht_pCitS,putative CitS from Halobacillus trueperi(AccessionNo.CDQ28507); Hs_pCitS,putative CitS from Halobacillus sp.BAB-2008(AccessionNo. ZP_20420905); Bsp_pCitS,putative CitS from Bacillus sp.SG-1(_01858 No.ZP); putative CitS from Bacillus halod...
Embodiment 2
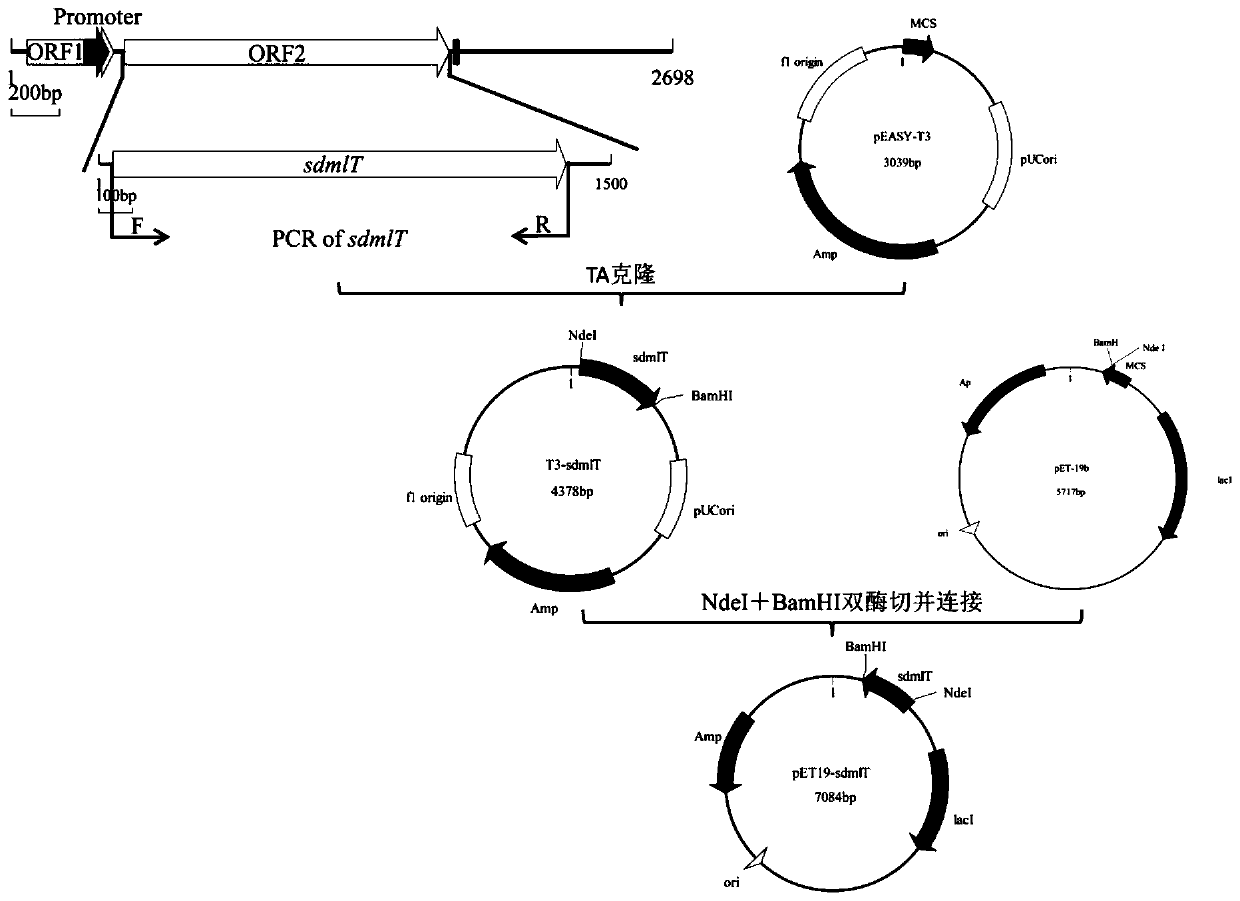
[0086] Example 2 Construction process of pET19-sdmlT prokaryotic expression vector; image 3 Shown
[0087] According to the sdmlT gene sequence, first use Primer5 software to design SEQID NO.3: sdmlT-F; SEQIDNO.4: sdmlT-R specific primers (as shown in Table 2), and introduce Nde I at the 5'and 3'ends of the gene. , BamH I two restriction sites, and then the sdmlT gene amplification (as shown in Table 3), the expected length of the amplified middle sequence fragment is about 1327bp.
[0088] Table 2 Sequences of primers constructed by Hz_sdmlT gene expression vector
[0089]
[0090] Table 3 PCR amplification reaction
[0091]
[0092] Add "A" to the end of the above PCR product and connect it to pEASY-T3 vector, then transfer it to Trans1-T1 competent cells, and perform blue-white spot screening to select subclones. Then treat the pEASY-T3-sdmlT with sdmlT gene with Nde I and BamH I, and do the same treatment on the expression vector pET19b at the same time, and recover and connect ...
Embodiment 3
[0099] Example 3 Identification of physiological function of sdmlT gene:
[0100] In order to identify the physiological function of the sdmlT gene, the present invention uses KNabc / pET19 and DH5α / pET19 as negative controls, and the subclones KNabc / pET19-sdmlT and DH5α / pET19-sdmlT are transferred to contain different salts at 1% inoculum. concentration( Figure 7 &8) Cultivate 24h in fresh LBK medium.
[0101] 1. Salt and alkali resistance test of KNabc / pET19-sdmlT
[0102] The salt-tolerance test of SdmlT in E.coli KNabc showed that the gene sdmlT can provide E.coli KNabc resistance to 0.4M NaCl, 5mM LiCl and alkaline pH (Figure 7), suggesting the gene sdmlT has salt and alkali resistance and may have Na + / H + Antiporter activity.
[0103] 2. Salt and alkali resistance test of DH5α / pET19-sdmlT:
[0104] The salt-tolerance test of SdmlT in wild-type E.coli DH5α shows that the gene sdmlT can increase the NaCl resistance of E.coli DH5α from 0.8M to 1.2M, and it also improves E.coli ( ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com