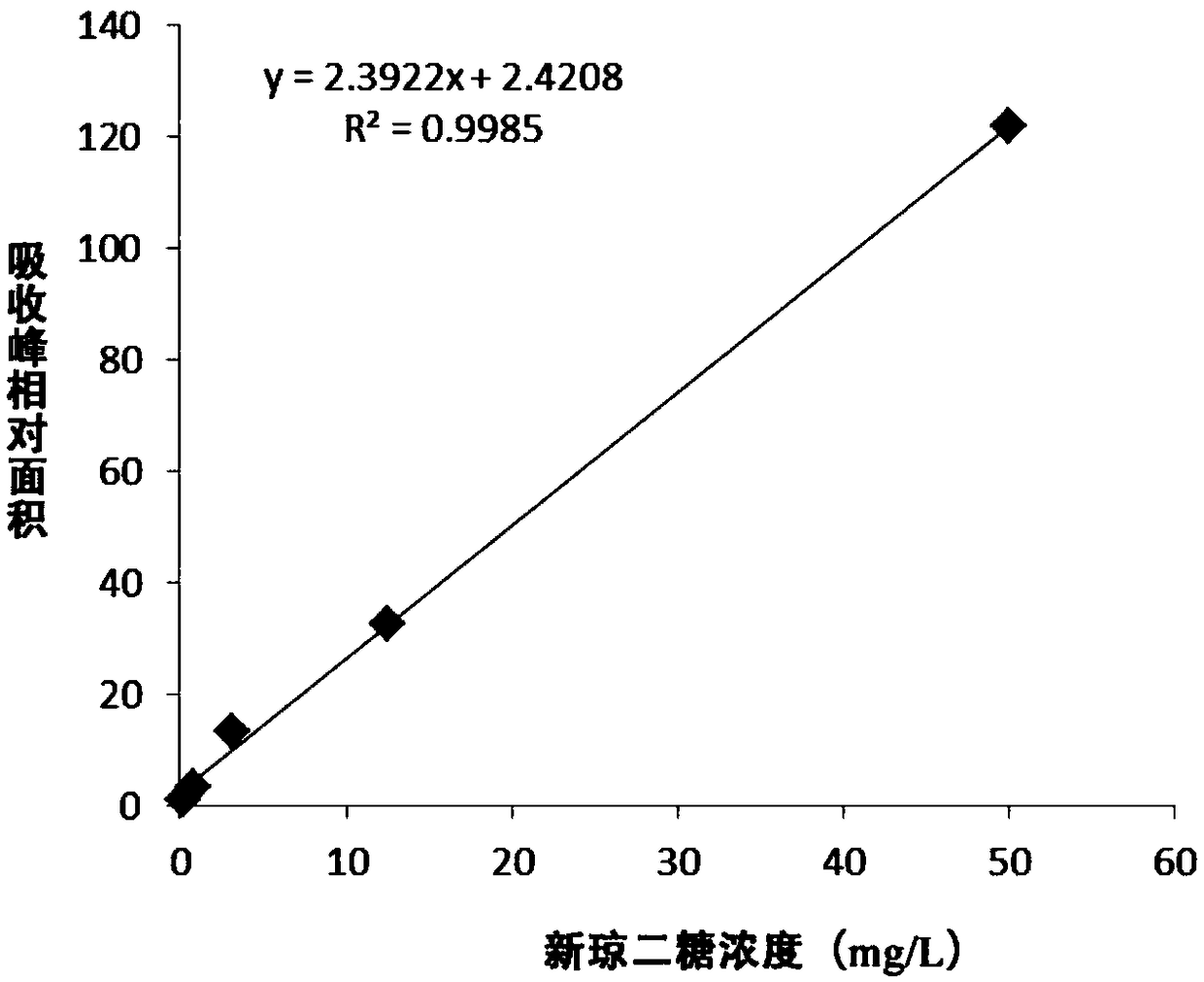
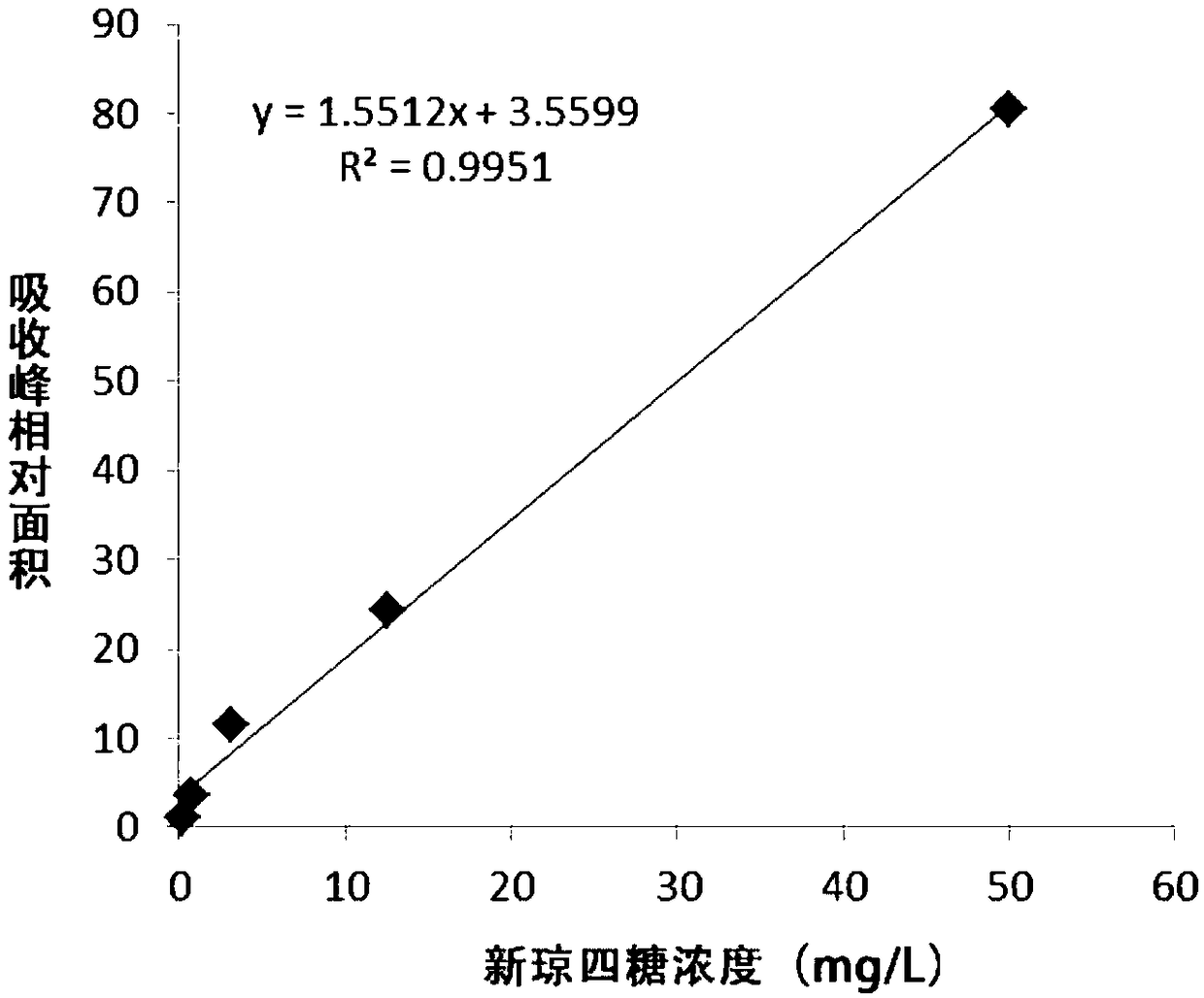
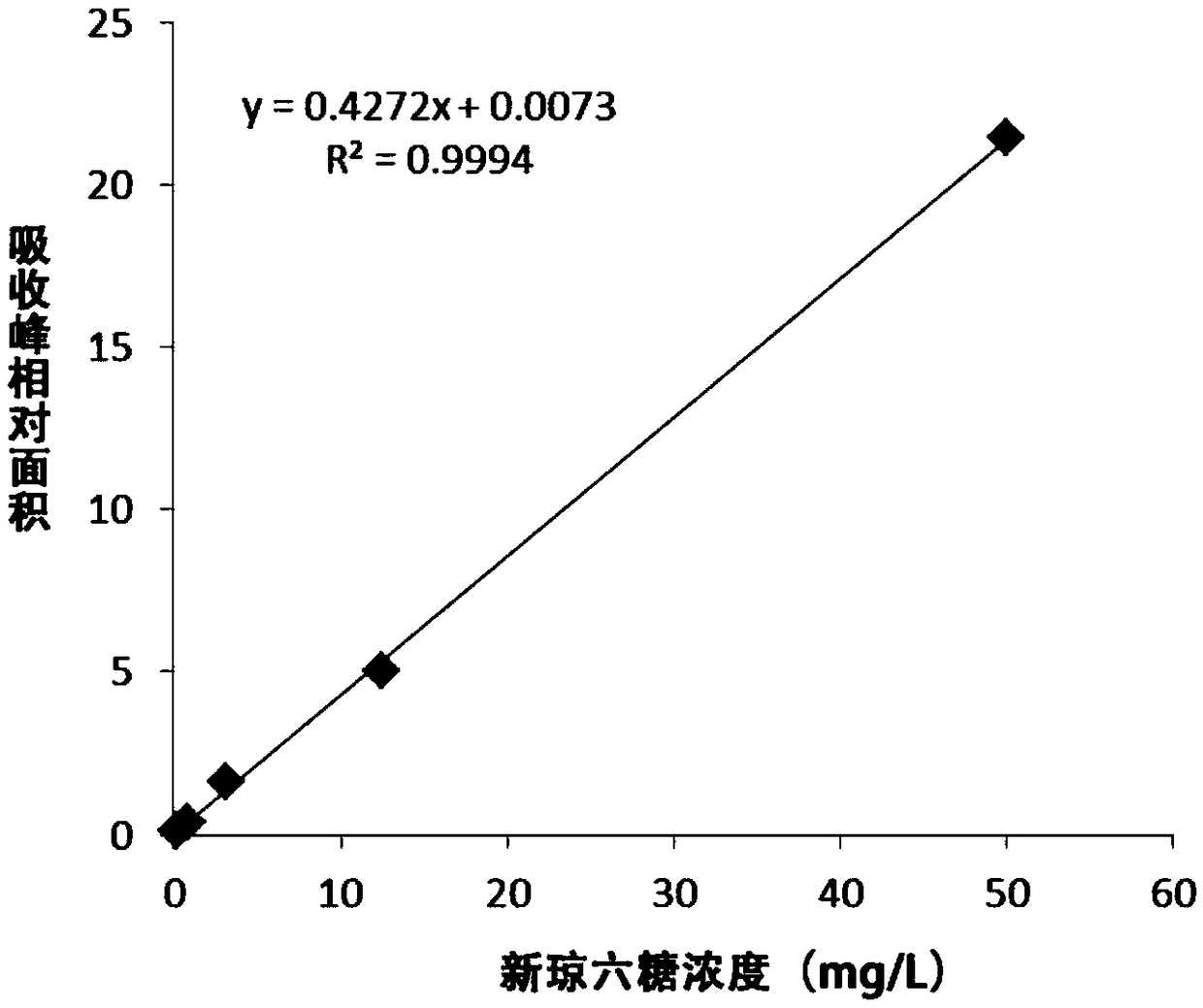
Method for producing neoagarobiose
A new technology of agarose and new agarose, which is applied in the field of enzyme engineering and bioengineering, can solve the problem of inability to determine the effective or main active components, large differences in the composition of agar degradation products, and the degree of polymerization of agar oligosaccharides. Great impact and other problems, to achieve the effect of improving uniformity, simple operation, and improving degradation ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1. Source and Enzyme Activity Analysis of Agarase Agaz306 and Agaz308
[0052] Our research group isolated a marine strain Flammeovirgapacifica WPAGA1 that can degrade agarose from deep sea sediments. Genome sequencing analysis obtained 13 β-agarases, and PCR technology was used to amplify the related agarase genes. The related agarase gene obtained was recovered by agarose gel electrophoresis, and then cloned into the expression vector pEASY-Blunt E2, and transferred into E.coli DH5α, and then sequenced and analyzed to verify the gene sequence. The correct positive clone was selected, cultured overnight at 37°C in LB medium containing 50 μg / mL ampicillin, the plasmid was extracted and transformed into the expression host E.coli BL21(DE3). The positive clones containing related recombinant plasmids were cultured in 50 mL SOC medium containing 50 μg / mL ampicillin at 37 °C until OD 600 When it is about 0.5, add 1 mM IPTG and induce at 16°C for 12 hours. After c...
Embodiment 2
[0055] Embodiment two, the preparation of recombinant plasmid
[0056] 1. Construction of recombinant plasmid pET-Sul1
[0057] (1) First, using primers Sulz308F and Sulz308R, the sulfatase Sulz308 was amplified from the genome of the strain Flammeovirga pacifica WPAGA1;
[0058] (2) Synthesize the signal peptide oligonucleotide chain PelB-Sulz308, the 18 bases at the end of the sequence are the sequence of sulfatase Sulz308;
[0059] (3) Using the above-mentioned Sulz308 gene sequence and signal peptide PelB-Sulz308 as a template, using primers PelB-Sulz308F and Sulz308RC to carry out PCR amplification to obtain the sulfatase Sulz308 sequence fused with the signal peptide PelB at the 5' end;
[0060] The PCR amplification system is as follows:
[0061]
[0062] The PCR reaction conditions are as follows:
[0063]
[0064] The second step to the fourth step reaction were repeated for 35 cycles.
[0065] (4) After recovering the PCR fragment, digest it with restrictio...
Embodiment 3
[0101] Embodiment 3, preparation and growth curve determination of engineering bacteria E.coli BL21 (pET-Sul1, pACY-NAB1)
[0102] (1) Prepare the E.coli BL21 strain containing the recombinant plasmid pACY-NAB1 into competent cells according to the method in the Molecular Cloning Experiment Guide.
[0103] (2) Transfer the recombinant plasmid pET-Sul1 into E.coli BL21 containing the recombinant plasmid pACY-NAB1 to obtain expression strains containing two recombinant plasmids pET-Sul1 and pACY-NAB1.
[0104] (3) Inoculate the recombinant E.coli BL21 containing two recombinant plasmids pET-Sul1 and pACY-NAB1 into 50 mL of SOCB medium containing 25 μg / mL chloramphenicol and kanamycin respectively; E.coli BL21 was inoculated in 50mL SOCA medium containing 25μg / mL chloramphenicol, cultured on a shaker at 37°C and 200rpm, until OD 600 When the temperature is 0.6, add IPTG with a final concentration of 0.1 mM, and continue to culture on a shaker at 24°C.
[0105] (4) E.coli BL21 c...
PUM
| Property | Measurement | Unit |
|---|---|---|
| degree of polymerization | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com