SNP Molecular Marker Combination for Rice Genotyping and Its Application
A whole-genome and rice technology, applied in the fields of molecular biology, genomics, bioinformatics and molecular plant breeding, can solve the problems of large-scale commercial breeding, cumbersome operation process, and small quantity, and achieve easy automatic detection , high flux, low cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
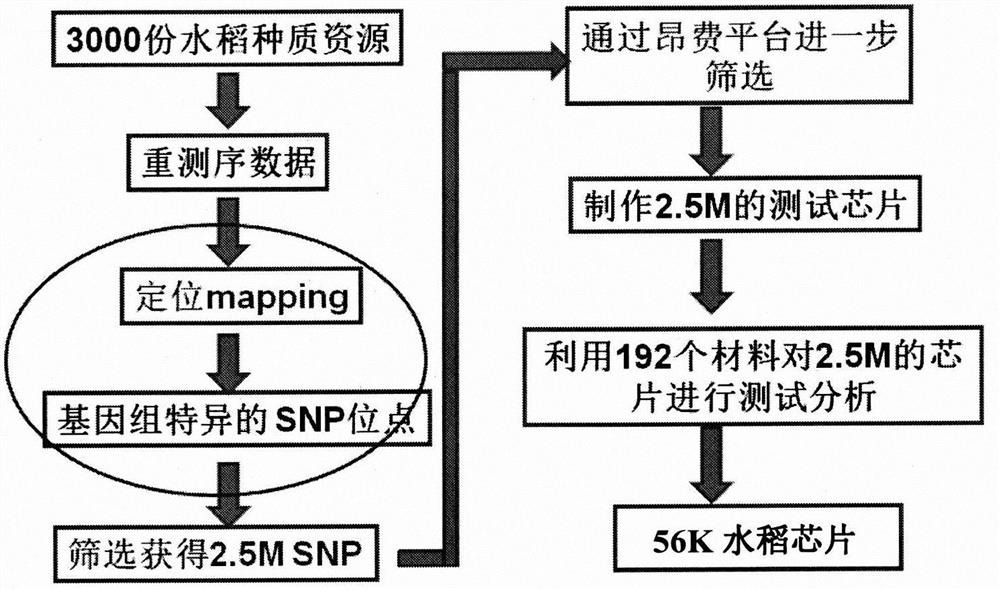
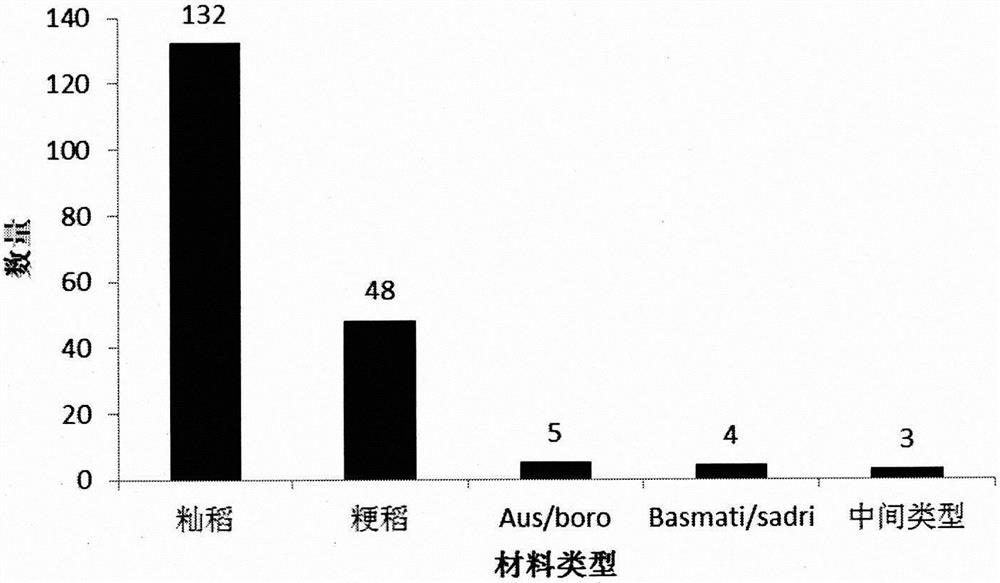
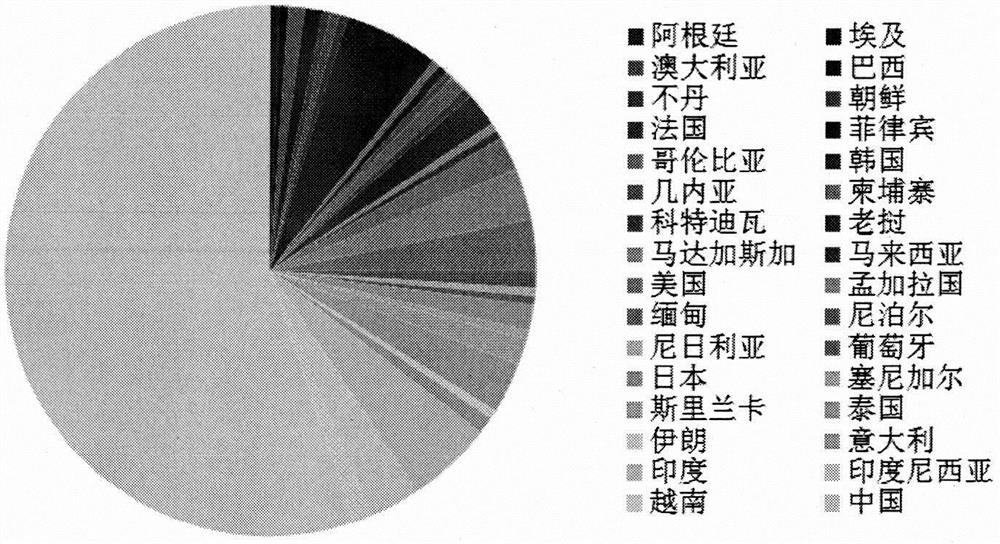
[0030] Example 1 Development of SNP Molecular Marker Combinations for Rice Genotyping
[0031] 1. Resequencing of 3024 rice germplasm resources and acquisition of SNP sites
[0032] 1. Library construction and sequencing of 3024 rice germplasm resources:
[0033] The DNA sequences of 24 random samples were added with 6 bp linker sequences and mixed together to construct a mixed index library. The linker sequences were used to distinguish the DNA sequences of each sample. The constructed library was subjected to PE90 sequencing on the HiSeq2000 machine, and at least 6 lanes were measured for each library to ensure that each sample could get enough data. Then split the data according to the linker sequence, and the read length of each sample after splitting is 83bp (83=90-6-1, 1 is the connecting base "T"). Finally, the reads contaminated by adapters and low-quality reads (base quality value ≤ 5) were removed to obtain high-quality cleanreads.
[0034] 2. Alignment and SNP / In...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com