A method for fast nucleic acid extraction
An extraction method and nucleic acid technology, applied in biochemical equipment and methods, DNA preparation, microbial measurement/inspection, etc., can solve the problems of complex extraction process and long time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] PCR results for the detection of Mycoplasma pneumoniae in nasopharyngeal swab samples from DNA lysates. Proceed as follows:
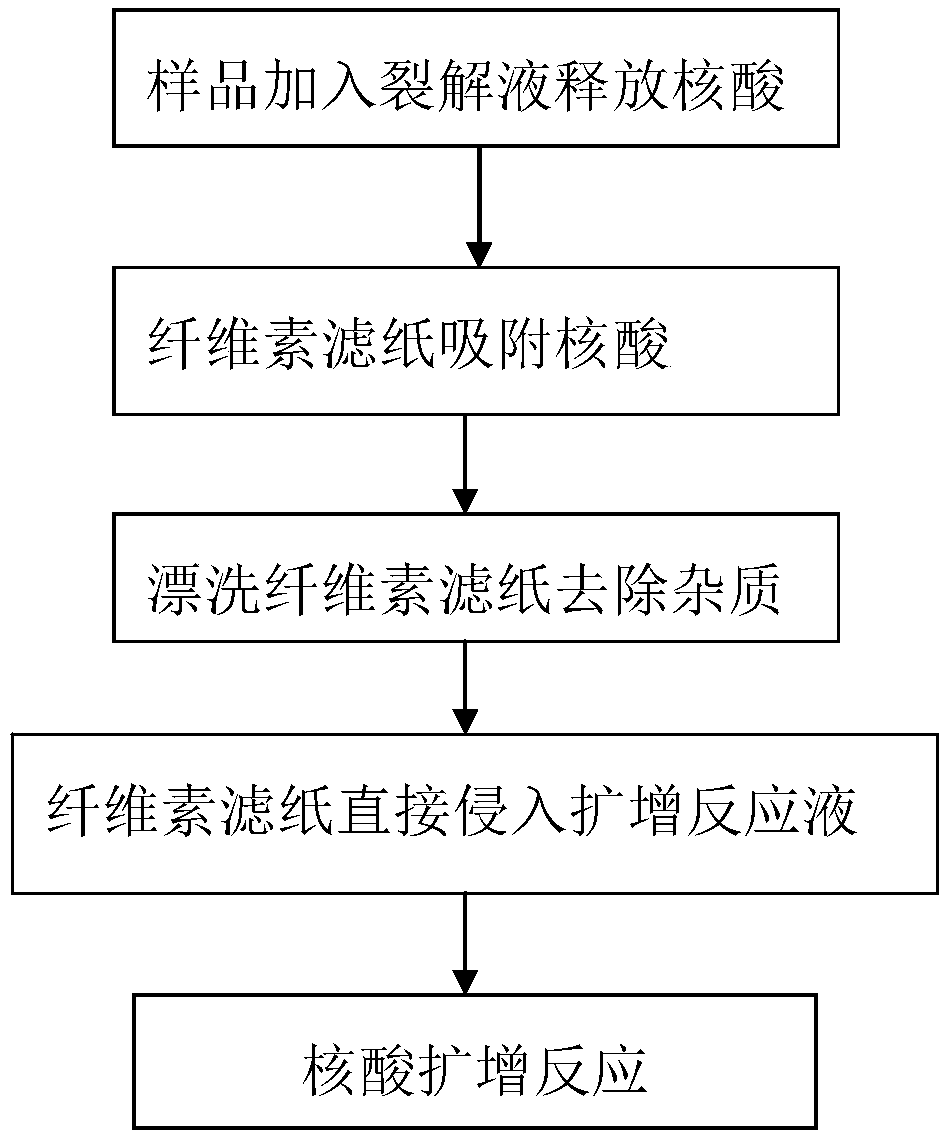
[0039] 1. Dip the nasopharyngeal swab with the sample directly into 1ml DNA lysis solution, 1ml DNA lysis solution is 1.5M guanidine hydrochloride, 50mM Tris (PH=8.0), 100mM NaCl, 5mM EDTA and 1% Tween20, and treat for 10 seconds .
[0040] 2. Immerse the cellulose filter paper in the DNA lysate in step 1 for 10 seconds.
[0041] 3. Immerse the cellulose filter paper obtained in the step 2 into the rinsing liquid, the rinsing liquid is 10 mM Tris pH=8.0 and 0.1% Tween-20, and treat for 10 seconds.
[0042] 4. Immerse the cellulose filter paper obtained in step 3 into the PCR reaction system for 10 seconds, or leave it in the reaction system.
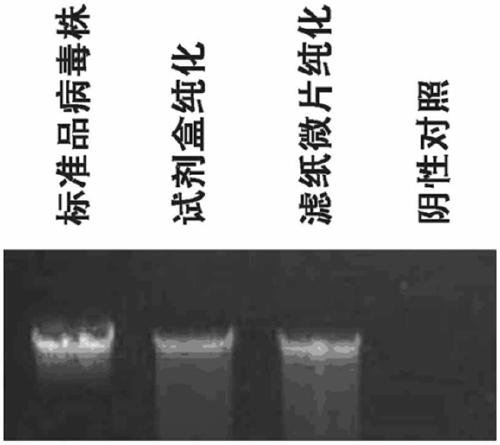
[0043] 5. PCR was amplified for 90 minutes. The same sample extracted from the standard plasmid and bacterial genomic DNA extraction kit (Beijing Tiangen) was used as a positive control, and the nasophar...
Embodiment 2
[0046] Detection of RPA results for influenza A in nasopharyngeal swab samples treated with RNA lysate. Proceed as follows:
[0047] 6. Immerse the nasopharyngeal swab with the sample directly into 1ml RNA lysis solution, 1ml RNA lysis solution is 4.0M guanidine isothiocyanate, 25mM sodium citrate, 0.5% (m / V) sodium lauryl creatine, 0.1mM β Mercaptoethanol, process for 10 seconds.
[0048] 7. Soak the cellulose filter paper in the RNA lysate obtained in step 6 for 10 seconds.
[0049] 8. Immerse the cellulose filter paper obtained in step 7 in the rinse solution, which is 10 mM Tris (pH=8.0) and 0.1% Tween-20, for 10 seconds.
[0050] 9. Immerse the cellulose filter paper obtained in step 8 into the RPA reaction system for 10 seconds, or leave it in the reaction system.
[0051] 10. RPA was amplified for 15 minutes. The same sample extracted from the standard virus strain and the virus RNA extraction kit (Shanghai Sangong) was used as a positive control, and the nasopharyng...
Embodiment 3
[0054] Detection of RPA results for dengue fever in RNA lysate-treated serum samples. Proceed as follows:
[0055] 11. Add 300ul of dengue fever patient serum sample to 1.2ml RNA lysate, the RNA lyse solution is 50μg / ml Proteinase K, 25mM sodium citrate, 0.5% (m / V) sodium laurate, 4.0M guanidine isothiocyanate and 0.1 mM β-mercaptoethanol,
[0056] 12. Treat with 50 μg / ml Proteinase K, 25 mM sodium citrate, 0.5% m / V sodium laurel creatine and 0.1 mM β-mercaptoethanol for 1 hour, then add 4.0 M guanidine isothiocyanate for 10 seconds.
[0057] 13. Soak the cellulose filter paper in the RNA lysate obtained in step 12 for 10 seconds.
[0058] 14. Immerse the cellulose filter paper obtained in step 13 into a rinsing buffer, and the rinsing solution is 10 mM Tris (pH=8.0) and 0.1% Tween-20 for 10 seconds.
[0059] 15. Immerse the cellulose filter paper obtained in step 14 into the RPA reaction system for 10 seconds, or leave it in the reaction system.
[0060] 16. RPA was ampli...
PUM
| Property | Measurement | Unit |
|---|---|---|
| area | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com