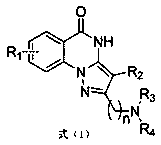
Pyrazole quinazolinone derivative serving as PARP (poly ADP-ribose polymerase) inhibitor and application of pyrazole quinazolinone derivative serving as PARP inhibitor
A technology of pyrazoloquinazolone and derivatives, which is applied in the field of pyrazoloquinazolone derivatives, which can solve problems such as no standard treatment plan, limited drug delivery efficiency, difficult onset of brain tumors, etc., and achieve oral Good bioabsorption, good application prospects, significant selective inhibitory effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1: Synthesis of compound I-1, prepared according to general scheme 1. The specific method is as follows:
[0050]
[0051] Step 1: Prepare SMb1: Add diethyl oxalate d1 (5 g) to a solution of sodium ethoxide (2.55 g, 37.7 mmol) in ethanol (50 ml), stir at 60°C for 0.5 h, and add propionitrile e1 (3 ml) Add to the mixture and heat to reflux for 7 h. The reactant was cooled to room temperature, the solid was collected by filtration, washed with ether, and naturally dried to obtain SMb1, which was directly used in the next reaction.
[0052] Step 2 Preparation c1: It can be prepared by General Method A and General Method B.
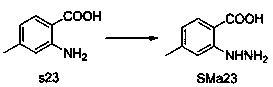
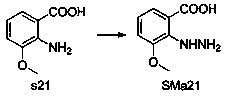
[0053] General method A: The raw material SMa1 is dissolved in acetic acid, SMb1 is added, the mixture is reacted in a microwave reactor at 150°C for 5-10 minutes, the precipitate is collected by filtration, washed with ether, and recrystallized with ethyl acetate / ethanol to obtain c1.
[0054] General method B: Suspend the raw materials SMa1 and SMb1...
Embodiment 2
[0058] Example 2: Preparation of I-2.
[0059] Dissolve I-1 (15 mmol) and methyl iodide (15 mmol) in ethanol, add sodium ethoxide, stir at 0℃~40℃ for 6h-10h, recover the solvent and perform silica gel column chromatography (ethyl acetate: petroleum ether 1 : 3) I-2 is separated. MS m / z (ESI): 243.1[M+1] + . 1 HNMR (400 MHz, DMSO-d6) δ 11.31 (s, 1H), 10.24 (s, 1H), 7.50-7.52 (m,1H), 7.81 (dd, 1H), 8.04 (d,1H), 8.09 (dd , 1H), 4.18 (s,2H), 3.23 (s,3H), 2.05 (s,3H).
Embodiment 3
[0060] Example 3: Preparation of I-3.
[0061] In the same manner as in Example 2, I-1 (10 mmol) and methyl iodide (30 mmol) were dissolved in ethanol, sodium ethoxide was added, and the mixture was stirred at reflux for 18 h-24 h. The solvent was recovered and subjected to silica gel column chromatography (ethyl acetate : Petroleum ether 1:5) I-3 is isolated. MS m / z(ESI): 257.1 [M+1] + . 1 HNMR (400 MHz, DMSO-d6) δ 11.46 (s, 1H), 7.51-7.55 (m,1H), 7.86 (dd, 1H), 8.03 (d,1H), 8.14 (dd, 1H), 3.65 (s ,2H), 2.06 (s,3H ), 2.18 (s,6H ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com