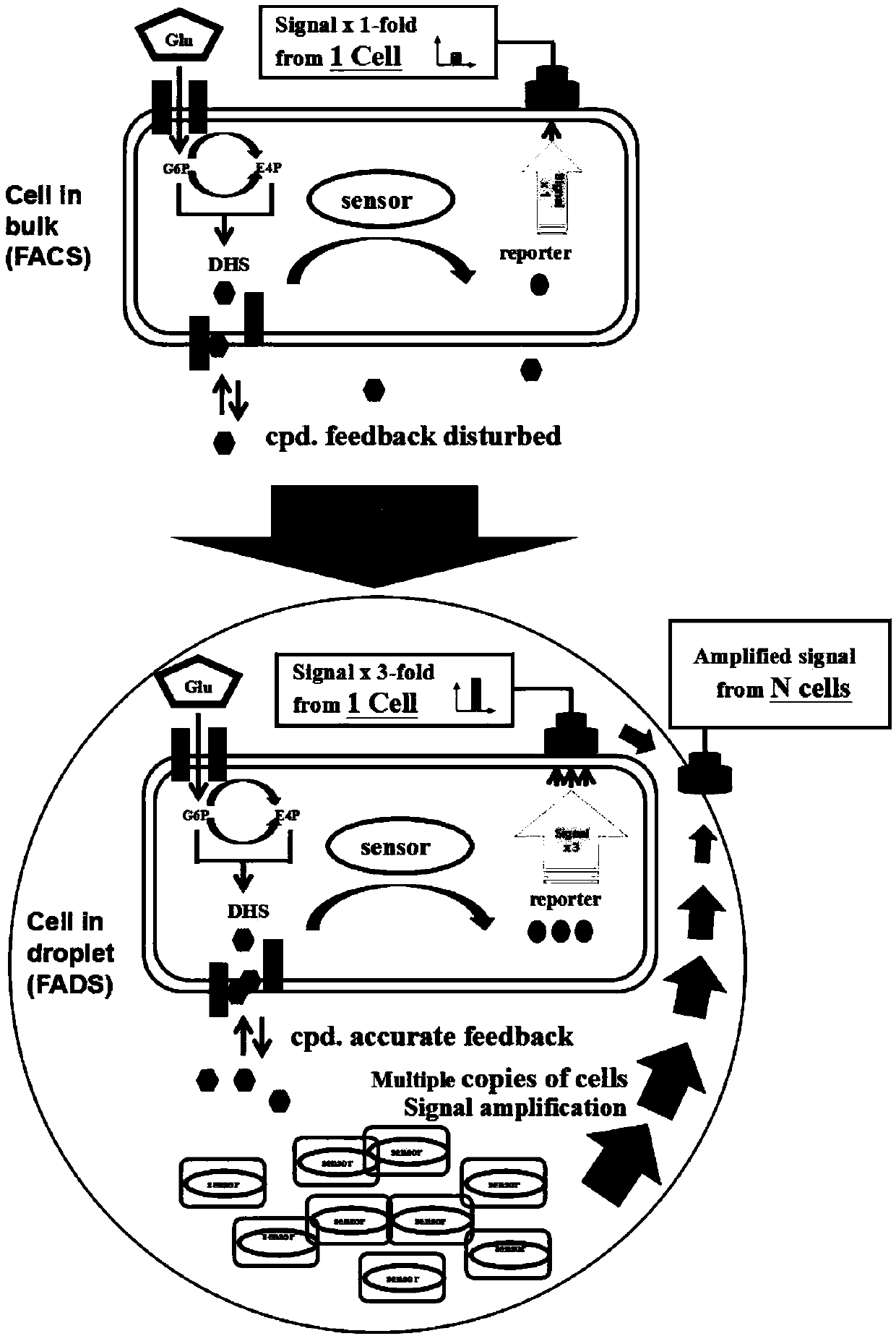
High-flux screening method based on biosensor
A biosensor and high-throughput technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve the problems of easy introduction of errors in fluorescent signals, difficult identification and sorting, background interference of fluorescent signals, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
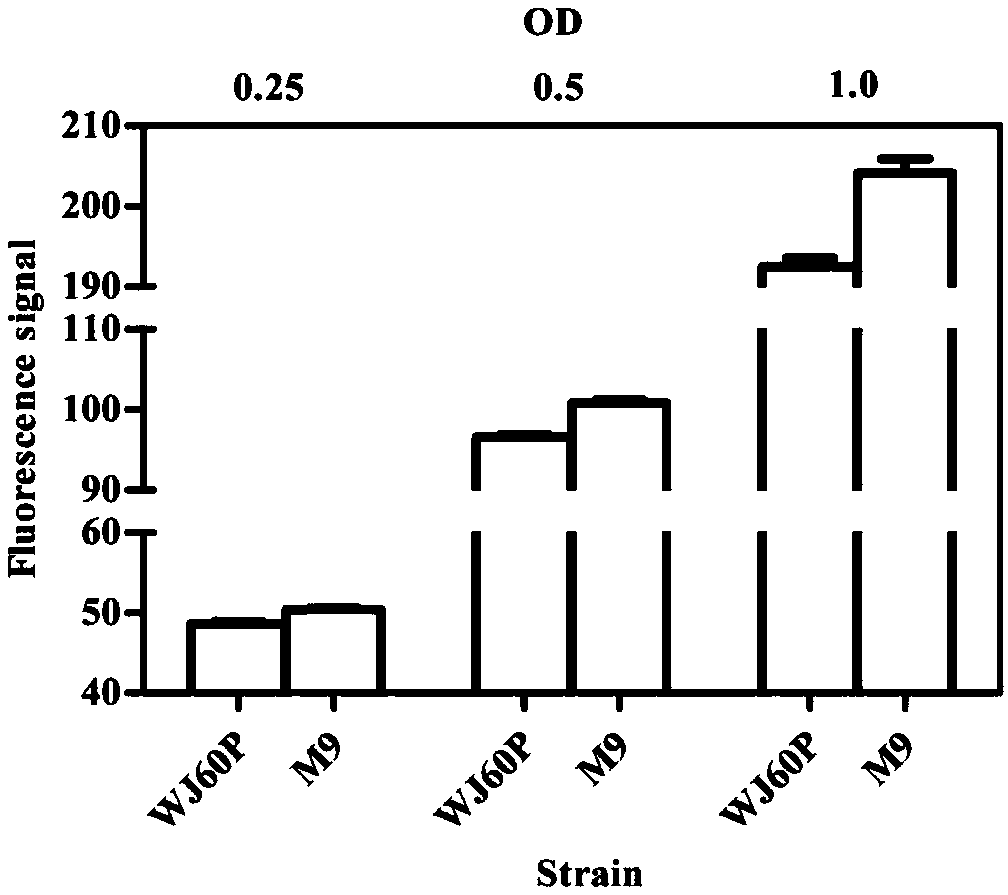
[0048] Example 1: Shake flask culture simulates microdroplet-embedded biosensor cell amplification signal difference experiment
[0049] Since the cells in the test tube or shake flask fermentation come from the same parental cell, which is consistent with the single cell embedding and propagation mechanism in the droplet, we use the shake flask to culture the single cell to simulate the micro droplet embedding biosensor cells To amplify the signal difference, the specific design is as follows: the strains WJ60P (weak) and M9 (strong) with differences in 3-dehydroshikimic acid (3-dehydroshikimic acid, DHS) synthesis ability were used as experimental strains, and the two strains were shaken under the same conditions. Fermentation culture, initial inoculation concentration: 0.1OD600, medium: NBS, temperature: 37°C, rotation speed: 220rpm, fermentation culture for 8 hours, sampling at fixed points, samples were washed twice with PBS and resuspended to 1OD600, and then serially dil...
Embodiment 2
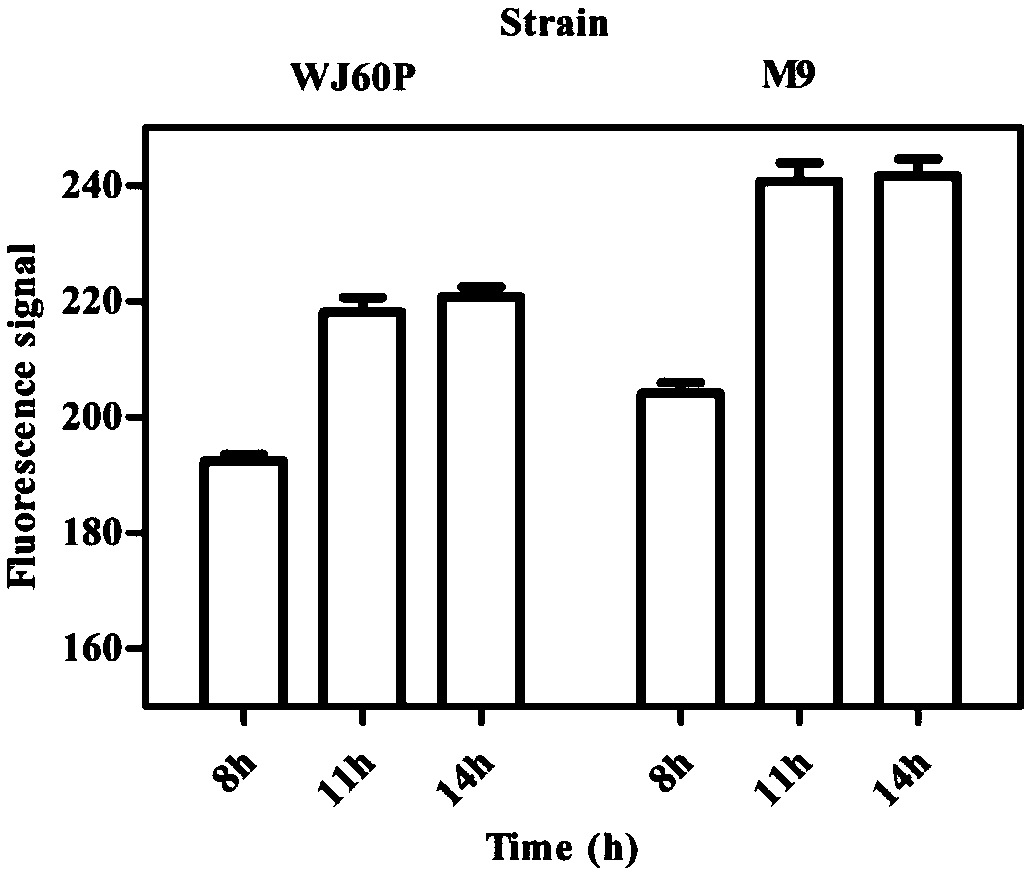
[0050] Example 2: Shake Flask Culture Simulated Microdroplet Encapsulation Biosensor Cell Feedback Mechanism Experiment
[0051] In order to simulate the cell feedback mechanism of microdroplet embedding biosensors, we used strains WJ60P (weak) and M9 (strong) with differential 3-dehydroshikimic acid synthesis ability as experimental strains, and carried out shake flask fermentation culture on the two strains under the same conditions , detect and observe the growth and fluorescence signal intensity changes of the two strains, compare the response fluorescence signal values of the biosensor in the two strains, and verify the effect of the extracellular feedback of the secreted small molecule compound on the sensing of the biosensor. In this experiment, the initial inoculation concentration: 0.1OD600, medium: NBS, temperature: 37°C, rotation speed: 220rpm, fermentation culture for 8h, 11h, 14h, sampling at fixed points, the samples were washed twice with PBS and resuspended to...
Embodiment 3
[0052] Example 3: Development and construction of a genotype biosensor that responds to the metabolic small molecule compound 3-dehydroshikimic acid
[0053] In order to construct a genotype biosensor that responds to the metabolic small molecule compound 3-dehydroshikimic acid, first, we took the transcriptome of the 3-dehydroshikimic acid-free strain E.coli ATCC 8739 as a reference, and rationally constructed the experimental group. The transcriptome samples of the 3-dehydroshikimic acid-producing engineering strain fermented for 12h and 20h were analyzed, combined with RT-qPCR verification, and the transcriptional regulator gene cusR positively correlated with the change of 3-dehydroshikimic acid concentration was excavated, and the gene cusR and its promoter are the core part, and the 15-amino acid short peptide with the coding sequence of 5'-ggtggtggtggttctggtggtggtggatccggtggcggtggttct-3' is used as the connecting sequence to connect the induction gene cusR with the promo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com