Chitinase CmChi6 gene and cloning expression and application thereof
A technology of chitinase and colloidal chitin, which is applied in the field of genetic engineering, can solve the problems of high energy consumption in the process, poor biological activity of the product, and low yield of the product, and achieve good repeatability, good protein expression effect, and high yield. The effect of degradation activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Chitinase cm Amplification of the Chi6 gene
[0046] The present invention designs primers based on the reported gene sequence of SYBC-H1 chitinase, and obtains SYBC-H1 chitinase by polymerase chain reaction (PCR) cm Conserved sequence of Chi6.
[0047] Using PCR technology, donated by Hao Zhikui of Jiangnan University Chitinolyticbacter meiyuanensis Genomic DNA of SYBC-H1 was used as a template, and primers were designed using Primer5.0 software, respectively: upstream primers Chi -F (SEQID No.2): 5'-CCCAAGCTTCTACTTTGCCGCCGGAAT-3' designed with NdeI restriction site; downstream primer Chi -R (SEQ ID No.3): 5'-CCCAAGCTTCTACTTTGCCGCCGGAAT-3' is designed with a HindIII restriction site.
[0048] PCR reaction system:
[0049]
[0050] PCR reaction conditions: 95°C pre-denaturation, 2min; 95°C denaturation for 20s, 52°C annealing, 20s, 72°C extension, 2min, a total of 30 cycles; 72°C extension, 5min, and finally 4°C incubation.
[0051] After the PCR r...
Embodiment 2
[0052] Example 2 Plasmid vector pColdI- Cmchi6 and the cloned strain pColdI- Cmchi6 - E. coli Preparation of Trans1T1
[0053] 1. Purify and recover the PCR product obtained in Example 1, using a TaKaRa company kit (TaKaRa DNALigation Kit);
[0054] 2、 Construction of plasmid vector pColdI- Cmchi6
[0055] The DNA sequence amplified by PCR and the pColdI vector were double-digested with the same restriction enzymes NdeI and HindIII, and the digested products were recovered and purified, and used T 4 DNA ligase was connected to obtain the plasmid vector pCooldI- Cmchi6 ;
[0056] 3. Construction of cloning strain pColdI- Cmchi6 - E. coli Trans1T1
[0057] The plasmid vector pColdI- Cmchi6 convert to E. coli Trans 1T1: (1) Take 20 μl of competent cells Trans 1T1 frozen and thawed on ice, and mix them gently in the above 10 μl connection reaction solution; (2) After standing on ice for 30 minutes, heat shock at 42°C for 45 seconds , and then quickly put it ...
Embodiment 3
[0062] Construction of recombinant expression strain pColdI- Cmchi6 - E. coli BL21(DE3)
[0063] 1. Strain cultivation: the constructed recombinant plasmid pColdI- Cmchi6 extracted from the cloning host and retransformed into E. coli. BL21(DE3) competent cells. The conversion operation was as the conversion step in the above-mentioned Example 2. Finally, 1-2 single colonies were picked and inoculated into 5ml LB medium containing ampicillin with a final concentration of 0.2%, and cultivated overnight at 37°C.
[0064] 2. Main culture: After 12 hours of culture, transfer to 100ml LB / Amp medium according to the inoculum size of 1%, and shake culture at 37°C until OD 600 =0.4-0.6.
[0065] 3. Induced expression: Add IPTG at a final concentration of 0.05mM to induce bacterial cells, and induce at 18°C and 200rpm for 20h at low temperature.
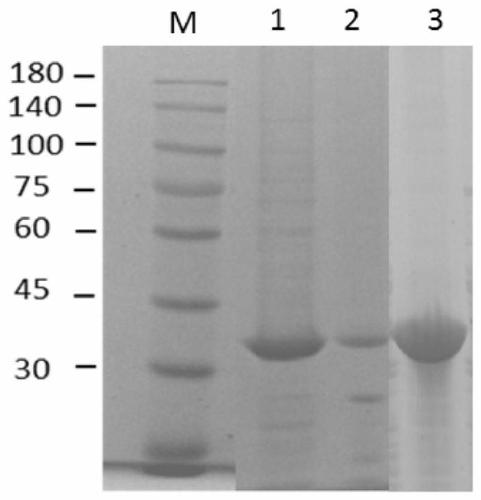
[0066] 4. Collect whole cells: Centrifuge at 4°C, 6000rpm for 8-10min to collect the bacteria, mix with 10ml PBS, and then ultr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com