Hibiscus esculentus reference gene EF-1 alpha and application thereof
An internal reference gene, EF-1 technology, applied in the field of molecular biology, to achieve the effect of improving detection efficiency, improving reliability and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1 Obtaining of internal reference gene EF-1α
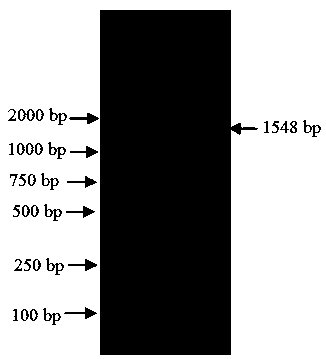
[0024] (1) Design a pair of primers according to the EF-1α gene of upland cotton (Gossypium raimondii) and Asian cotton (Gossypium arboreum L.) on GenBank, and use the clustalx software to perform sequence alignment on the designed primer pair, and the platinum Shang Biotechnology (Shanghai) Co., Ltd. synthesized the primer pair, specifically, the primer pair (as shown in SEQ ID NO.1, 2):
[0025] Forward primer 5'-TTCCGAGTTCTATAATGGGTAA-3',
[0026] Reverse primer 5'-ACGCCACAGCTTATTTCTTC-3'.
[0027] (2) DNA extraction: Weigh about 0.1 g of okra leaves, add liquid nitrogen to quickly and fully grind to powder (add 0.1 g of PVP), transfer the powder to a 1.5 mL centrifuge tube; add 600 μL to each tube and preheat to 65 2×CTAB extraction buffer at ℃, add 7 μL of 2-mercaptoethanol at the same time, mix thoroughly, put in a water bath at 65℃ for 30 min, and invert several times; take out the centrifuge tube, cool to ...
Embodiment 2
[0032] Example 2 Real-time fluorescent quantitative PCR design and routine PCR detection
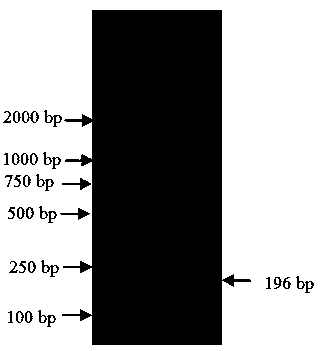
[0033] Based on the nucleotide sequence of the internal reference gene obtained in Example 1, using Primer Premier5.0 software, and following the principles of real-time fluorescent quantitative PCR primer design, a pair of fluorescent quantitative specific primers were designed, and the amplified fragment was 196 bp. The pair of fluorescent quantitative specific primers are the real-time fluorescent quantitative PCR primers (as shown in SEQ ID NO: 4, 5):
[0034] Forward primer 5'-GCTGCCAAGAAGAAATAAGC-3',
[0035] Reverse primer 5'-ATGAAGCAAACCTCGACACT-3'.
[0036] The total RNA of okra was extracted, and the first strand of cDNA was synthesized according to the method of PrimeScriptTM 1st Strand cDNA Synthesis Kit, that is, the RNA was reverse transcribed into cDNA; then the obtained cDNA was used as a template and real-time fluorescent quantitative PCR primers were used as primer pai...
Embodiment 3
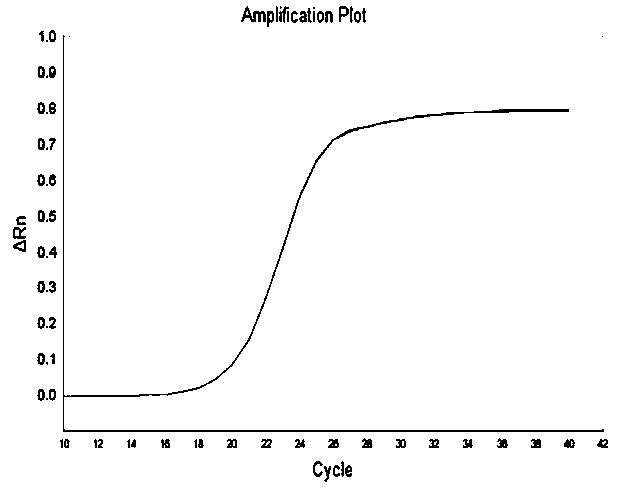
[0040] Example 3 Real-time fluorescent quantitative PCR primer verification
[0041] Extract the total RNA of okra, and synthesize the first strand of cDNA according to the method of PrimeScriptTM 1st Strand cDNA Synthesis Kit, that is, reverse transcribe the RNA into cDNA; then use the obtained cDNA as a template, follow the instructions of Power SYBR®Green PCR Master Mix in ABI7500 in real time The PCR reaction was carried out on the quantitative PCR instrument, and the reaction system and reaction procedure of the PCR reaction were as follows:
[0042] The reaction system is: the total volume of the reaction system is 25 μL, 12.5 μL Power SYBR® Green PCR MasterMix, 1 μL template, 0.5 μL of the forward primer of the real-time fluorescence quantitative PCR primer in Example 2 (concentration is 10 μmol / L), implement The reverse primer of the real-time fluorescent quantitative PCR primer in Example 2 was 0.5 μL (concentration: 10 μmol / L), and distilled water was added to make t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com