Application of recombinant escherichia coli with overexpression of fimH gene in fermentation production of amino acid
A technology of recombinant Escherichia coli and Escherichia coli, which is applied in the field of amino acid fermentation, can solve the problems of slow rate and achieve the effect of increasing production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
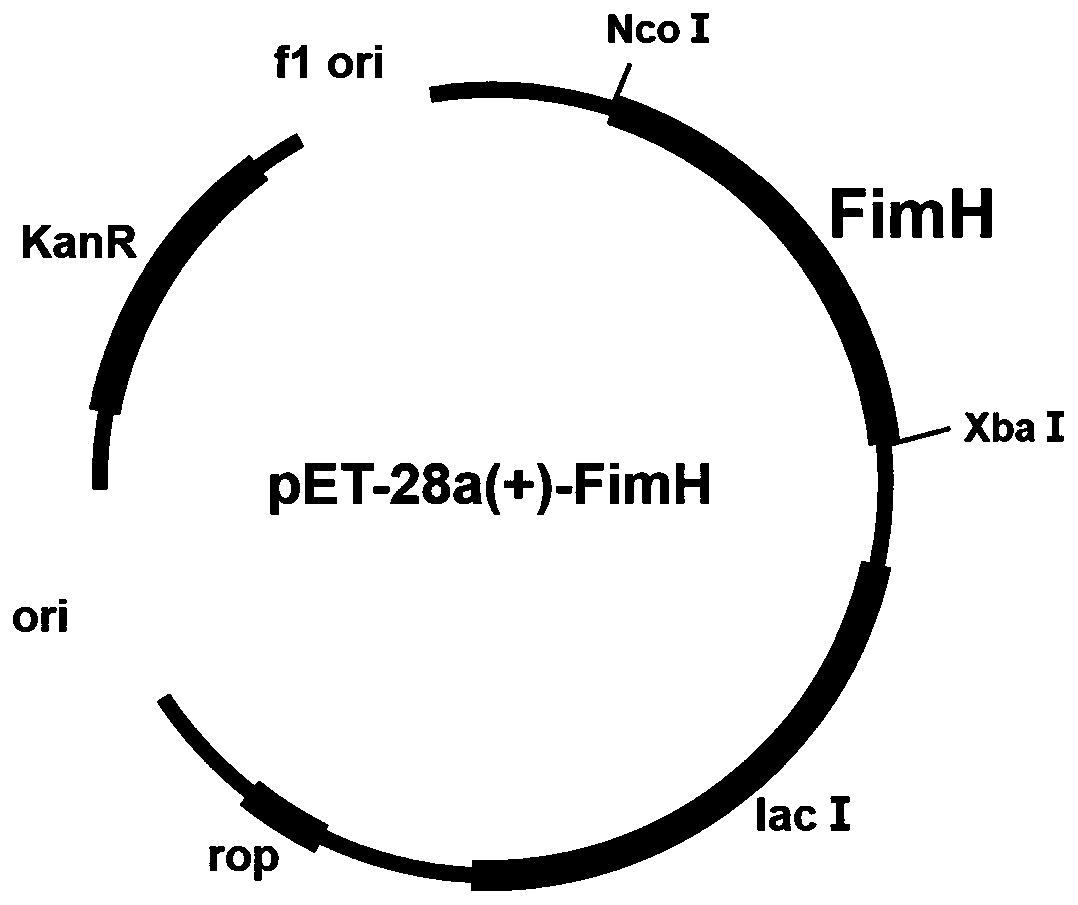
[0022] Embodiment 1 constructs the recombinant escherichia coli that overexpresses fimH gene
[0023] 1. Amplification of the target gene fimH: the gene fimH was amplified using the extracted Escherichia coli CIBTS1688 genome as a template. According to the fimH gene sequence of Escherichia coli str.K-12substr.MG1655 on NCBI, primers used for amplification were designed by snapgene software, wherein the primers included Xba I and Nco I restriction enzyme sites for subsequent connection with plasmid pET28a. The primers were synthesized by Jinweizhi Company in Suzhou City, Jiangsu Province.
[0024] Primer name and sequence:
[0025] fimH-F: GGATAACAATTCCCCTCTAGAATGAAACGAGTTATTACCCTGTTTG
[0026] fimH-R: GATGATGGCTGCTGCCCATGGTTATTGATAAACAAAAGTCACGCCA
[0027] The PCR reaction system is shown in Table 1.
[0028] Table 1 Target gene fimH amplification PCR system
[0029] components Volume (μL) Manufacturer 10×PCR buffer 25 μL TOYOBO Corporation dN...
Embodiment 2
[0044] Embodiment 2 constructs the recombinant escherichia coli that knocks out the fimH gene
[0045]1. Extract the knockout plasmids pKD46 and pKD4: inoculate Escherichia coli DH5α (purchased from Miaoling Plasmid Platform) Glycerol with plasmid pKD46 in 5 mL of LB liquid medium (containing 100 μg / mL ampicillin), and culture at 30°C Overnight; collect the cells in a 2.0 mL centrifuge tube, centrifuge at 10,000 rpm for 2 min, and discard the supernatant; follow the steps in the instructions of the AxyPrep Plasmid DNA Mini Kit from Corning Life Sciences to extract the plasmid pKD46. The extracted plasmids were stored at -20°C.
[0046] The formula of LB liquid medium (containing 100 μg / mL ampicillin) is as follows: yeast powder 5 g / L, peptone 10 g / L, sodium chloride 10 g / L, ampicillin 100 μg / mL.
[0047] Inoculate Escherichia coli DH5α (purchased from Miaoling Plasmid Platform) with plasmid pKD4 into 5 mL of LB liquid medium (containing 50 μg / mL kanamycin) and culture overnig...
Embodiment 3
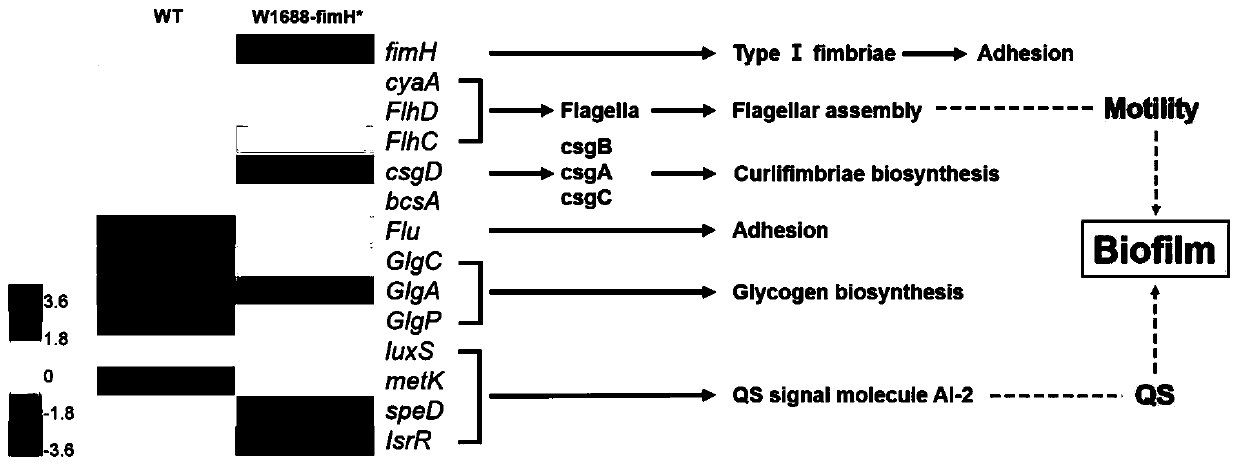
[0059] Example 3 Gene-wide transcriptome analysis of the original strain CIBTS1688, the knockout strain CIBTS1688-ΔfimH and the overexpression strain CIBTS1688-fimH*
[0060] 1. Activation: Take CIBTS1688, CIBTS1688-ΔfimH and CIBTS1688-fimH* out of the -80°C refrigerator, prepare 5 mL of LB medium in a test tube with an inoculum size of 30 μL, and culture them in a shaker at 37°C for 12 hours at a speed of 200 rpm.
[0061] 2. Fermentation: Transfer CIBTS1688, CIBTS1688-ΔfimH and CIBTS1688-fimH* to the fermentation medium and ferment for 30 hours at 37°C respectively, then take 2mL of the fermentation broth and put it into a 2mL centrifuge tube, and quickly put it into the pre-startup setting to 4 Centrifuge in a centrifuge at 12000rpm for 2min, discard the supernatant, immediately place the centrifuge tube in liquid nitrogen for 3min, take it out and put it in a -80℃ refrigerator for use. Three parallels were prepared for each group of samples.
[0062] 3. The transcriptome ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com