A method for detecting ochratoxin a using fluorescence anisotropy technique
An ochratoxin and anisotropic technology, applied in the field of detection of ochratoxin A, can solve the problems of high antibody preparation cost, poor antibody stability, cumbersome and time-consuming operation, etc., and achieve easy label introduction and high-throughput analysis , convenient storage and transportation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Embodiment 1, the preparation of nucleic acid aptamer and complementary nucleic acid sequence
[0059] 1. Preparation of nucleic acid aptamers
[0060] A nucleic acid aptamer (as shown in sequence 1 of the sequence listing) that can specifically bind ochratoxin A was artificially synthesized, and the 5' end of the nucleic acid aptamer was labeled with fluorescein (FAM).
[0061] 2. Screening and preparation of complementary nucleic acid sequences
[0062] For the nucleic acid aptamer prepared in step 1, design the following single-stranded DNA molecules that can complementarily bind to the nucleic acid aptamer:
[0063] Ⅰ: 5'-CCA CCC ACA CCC GAT C-3' (sequence 2 of the sequence listing);
[0064] Ⅱ: 5'-CAC CCA CAC CCG ATC-3' (sequence 3 of the sequence listing);
[0065] Ⅲ: 5'-ACC CAC ACC CGA TC-3' (sequence 4 of the sequence listing);
[0066] Ⅳ: 5'-CCC ACA CCC GAT C-3' (sequence 5 of the sequence listing);
[0067] V: 5'-CCA CAC CCG ATC-3' (sequence 6 of the sequ...
Embodiment 2
[0074] Example 2. Establishment of a method for detecting ochratoxin A using fluorescein-labeled nucleic acid aptamers and streptavidin-labeled single-stranded DNA molecules
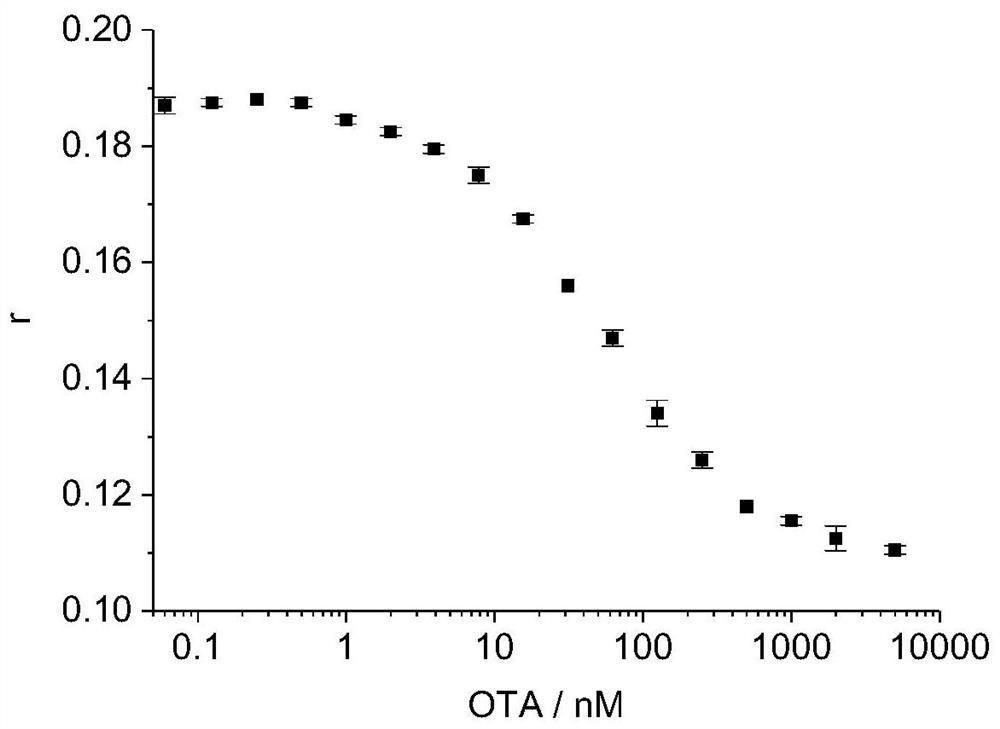
[0075] 1. Mix the FAM-labeled aptamer prepared in Example 1, the streptavidin-labeled single-stranded DNA molecule and ochratoxin A in 100 μl of reaction buffer solution, and incubate at 25° C. for 10 minutes. The concentration of the FAM-labeled nucleic acid aptamer in the reaction system is 5nM, the concentration of the streptavidin-labeled single-stranded DNA molecule in the reaction system is 10nM, and the ochratoxin A in the reaction system is set at different concentrations (per Each concentration set 2 replicates). At the same time, set a blank control without adding the sample to be tested.
[0076] 2. After the reaction in step 1, the fluorescence anisotropy (fluorescence polarization) value was measured with a multi-functional microplate reader (Synergy H1 Microplate reader, Biotek, USA), the ...
Embodiment 3
[0080] Embodiment 3, the impact of streptavidin labeling on detection sensitivity
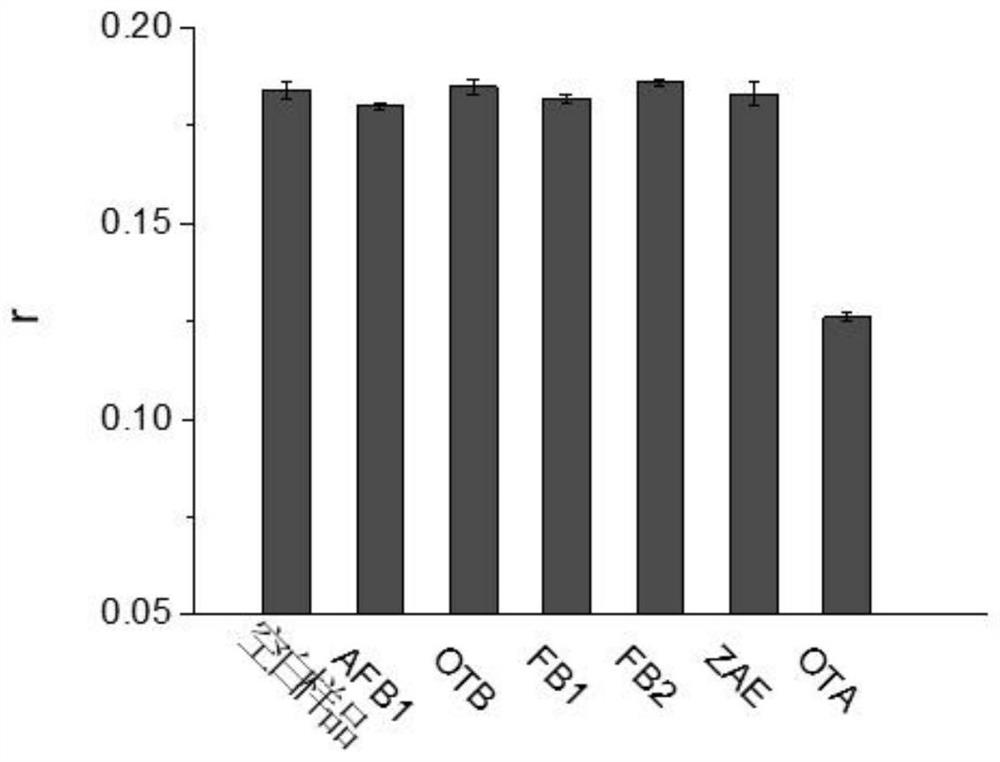
[0081] 1. The FAM-labeled nucleic acid aptamer prepared in Example 1, only the single-stranded DNA molecule shown in sequence 5 labeled with biotin (not labeled with streptavidin) was reacted with ochratoxin A in 100 μl Mix in buffer solution and incubate at 25°C for 10 min. The concentration of the FAM-labeled nucleic acid aptamer in the reaction system was 5 nM, and the concentration of only biotin-labeled single-stranded DNA molecules (not labeled with streptavidin) in the reaction system was 10 nM. The concentration of Aspergillus toxin A was 200 nM. At the same time, set a blank control without adding the sample to be tested.
[0082] 2. After the reaction in step 1, the fluorescence anisotropy (fluorescence polarization) value was measured with a multi-functional microplate reader (Synergy H1 Microplate reader, Biotek, USA), the excitation wavelength was 485nm, and the emission waveleng...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com