Application of RBP1 gene in sow ovarian granular cells
A technology of granulosa cells and sows, which is applied in the field of cell engineering and genetic engineering, and can solve the problems that the relationship between RBP1 gene and the growth and development of sow ovary granulosa cells has not been reported.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
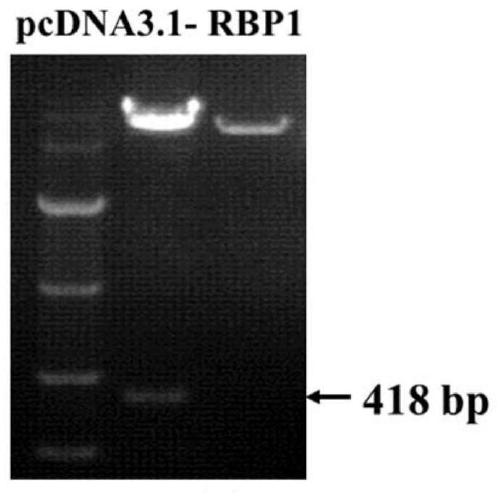
Embodiment 1
[0068] Example 1 ChIP-Seq screening of differential gene RBP1
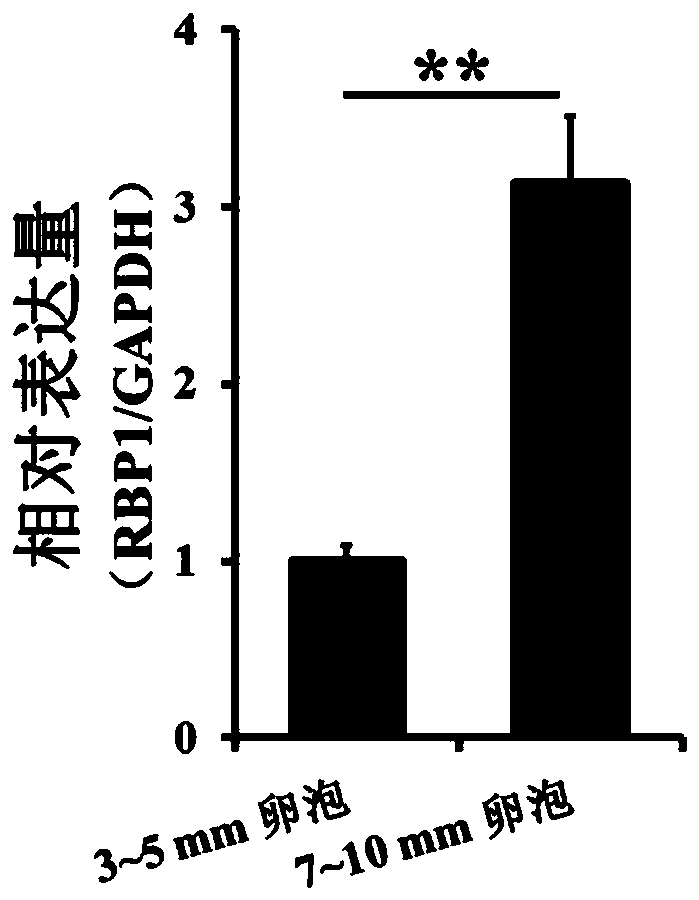
[0069] (1) Select two follicles with a diameter of 3-5 and 7-10mm in the sow ovary, formaldehyde cross-links the entire tissue, that is, connects the target protein with chromatin, separates the genomic DNA, and breaks the DNA by ultrasound; add the target protein The specific antibody anti-H3K4me3 (SANTA CRUZ) forms an immunoprecipitation complex; decrosslinking and purifying the DNA obtains a DNA sample of chromatin immunoprecipitation.
[0070] (2) DNA samples were sent to Shanghai Kangcheng Bioengineering Co., Ltd. for gene sequencing. The basic process of sequencing:
[0071] ①Sample quality assessment, using Quant-iT TM The dsDNA High-Sensitivity (HS) Assay Kit (Invitrogen) determined the purity and concentration of DNA samples.
[0072] ② Sequence library preparation, use TruSeq Nano DNA Sample Prep Kit (FC-121-4002, Illumina) to perform end repair, tail-end ligation and adapter ligation on DNA samples; ...
Embodiment 2
[0078] Example 2 RNA extraction, quality detection and reverse transcription
[0079] (1) RNA extraction:
[0080] ① Sample digestion: extract RNA from tissue samples, take 50-100 mg of tissue samples, cut them up quickly on ice or repeatedly homogenize with a homogenizer, and add Trizol (50-100 mg / ml). Extract RNA from cells without digesting cells, directly add Trizol (10cm 2 / mL).
[0081] ② Place the tissue or cell sample on ice for 10-15 minutes, centrifuge at 12,000 g at 4°C for 5 minutes, and transfer the RNA-containing supernatant to a new RNase-free tube.
[0082] ③Add chloroform (the volume ratio of chloroform:Trizol is 1:5), shake vigorously, place at room temperature for 5 minutes, centrifuge at 12,000g at 4°C for 15 minutes, and carefully transfer the upper aqueous phase to a new RNase-free tube.
[0083] ④Add isopropanol (volume ratio of isopropanol:Trizol is 1:2), mix by inverting slightly, place at room temperature for 10min, centrifuge at 12,000g for 15min ...
Embodiment 3
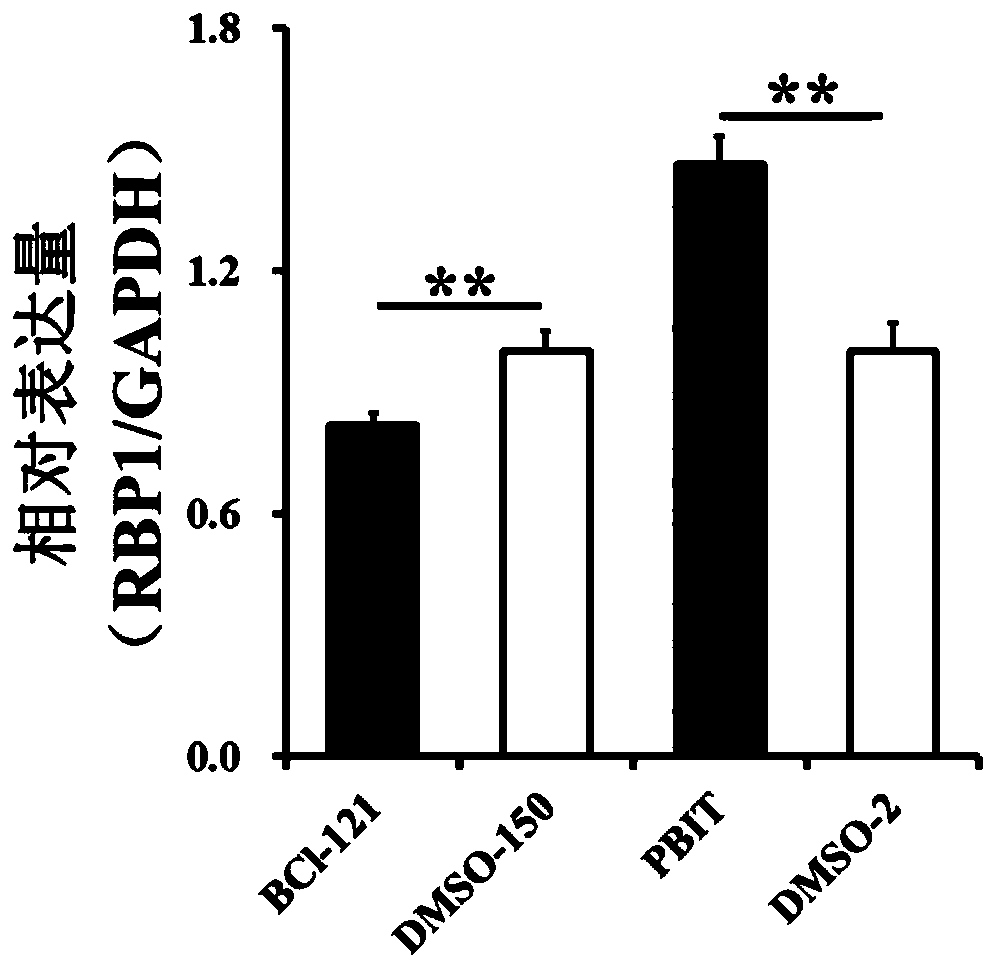
[0095] Example 3 qRT-PCR
[0096] The qRT-PCR detection of the genes in the present invention uses the Maxima SYBR Green qPCR Master Mix (2X) kit (Thermo Scientific). In the experiment, the comparative Ct value method was used to detect the content of the gene in the sample, and the specific calculation formula was as follows:
[0097] Relative gene expression = 2-{-〈﹙Ct value of the target gene in the control group﹚-﹙Ct value of the internal reference gene in the control group﹚>}
[0098] The detection gene uses GAPDH as an internal reference, and the qRT-PCR primers used in the present invention are:
[0099] qRT-PCR-RBP1 Forward: 5′-TAGGGTTCATTGTGGTCAGTGT-3′;
[0100] Reverse: 5'-CAATTTTGCGCAAGGCCACA-3';
[0101] qRT-PCR-GAPDH Forward: 5′-GGACTCATGACCACGGTCCAT-3′;
[0102] Reverse: 5'-TCAGATCCACAACCGACACGT-3'.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
| Diameter | aaaaa | aaaaa |
| Diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com