Application of high-temperature resistance Cas protein and detection system and kit for target nucleic acid molecule
A target nucleic acid, kit technology, applied in biochemical equipment and methods, microbial determination/inspection, hydrolase, etc., can solve problems affecting Toehold detection and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
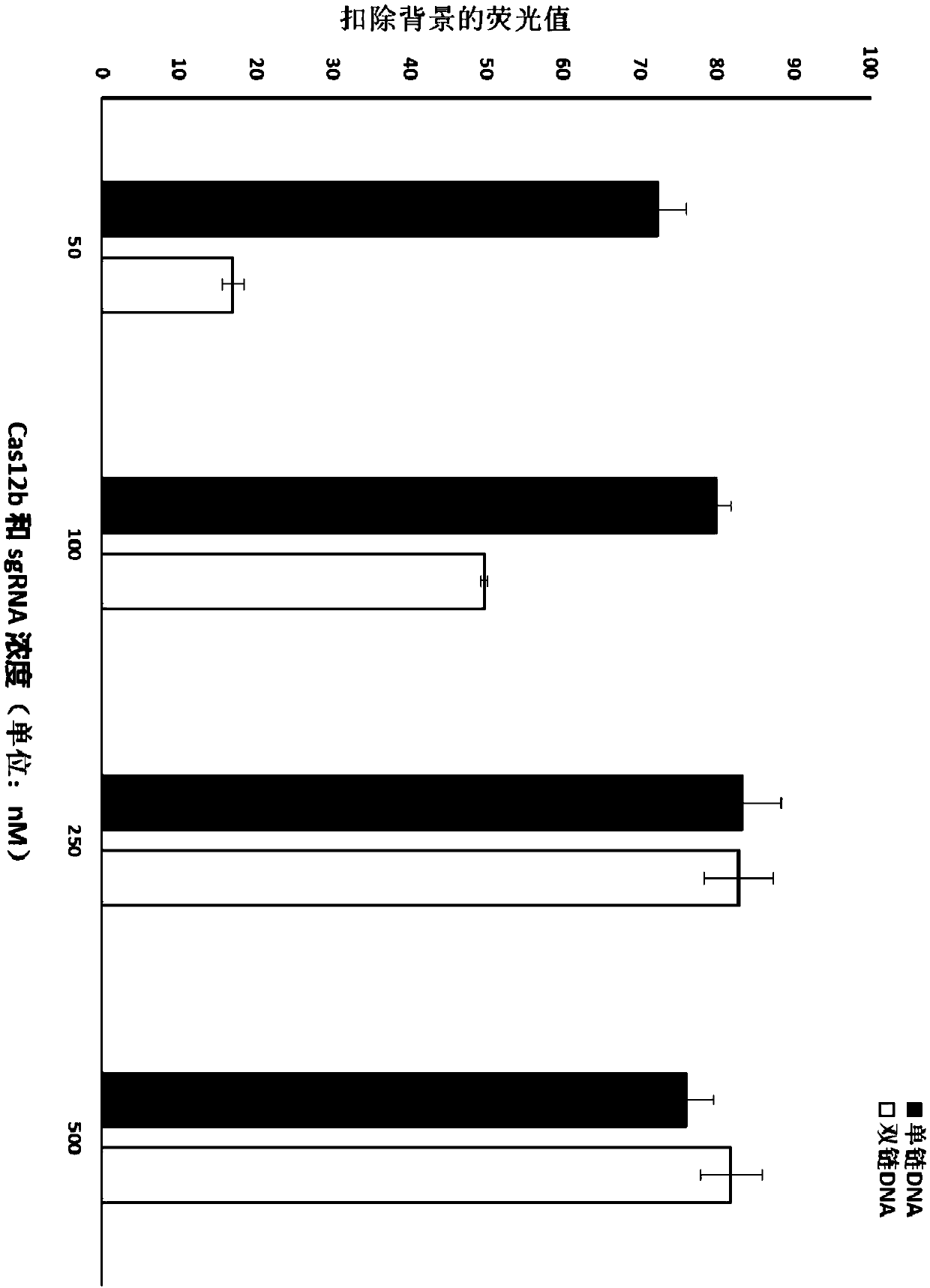
[0294] Embodiment 1Cas12b protein detects the target of double-stranded DNA (dsDNA) or single-stranded DNA (ssDNA)
[0295] Select single-stranded DNA or double-stranded DNA (target T1) as the target sequence, and test the response value of different concentrations of Cas12b protein and sgRNA to its detection.
[0296] 1. Preparation of guide RNA (sgRNA)
[0297] First construct the plasmid pUC18-sgRNA-T1 with pUC18 as the plasmid backbone, which inserts the T7 promoter and the template DNA sequence for transcription of sgRNA on pUC18 (Note: the sgRNA transcribed from this template targets a sequence called T1). The method is to use the pUC18 plasmid as a template, pUC18-1-F and pUC18-1-R as primers, first perform a round of PCR, T4DNA Ligase ligates the PCR product, transforms it into DH10b, and obtains a correct clone by sequencing, which is called pUC18- sgRNA-T1-pre. Then use pUC18-sgRNA-T1-pre as template, pUC18-2-F and pUC18-2-R as primers, carry out the second round ...
Embodiment 2
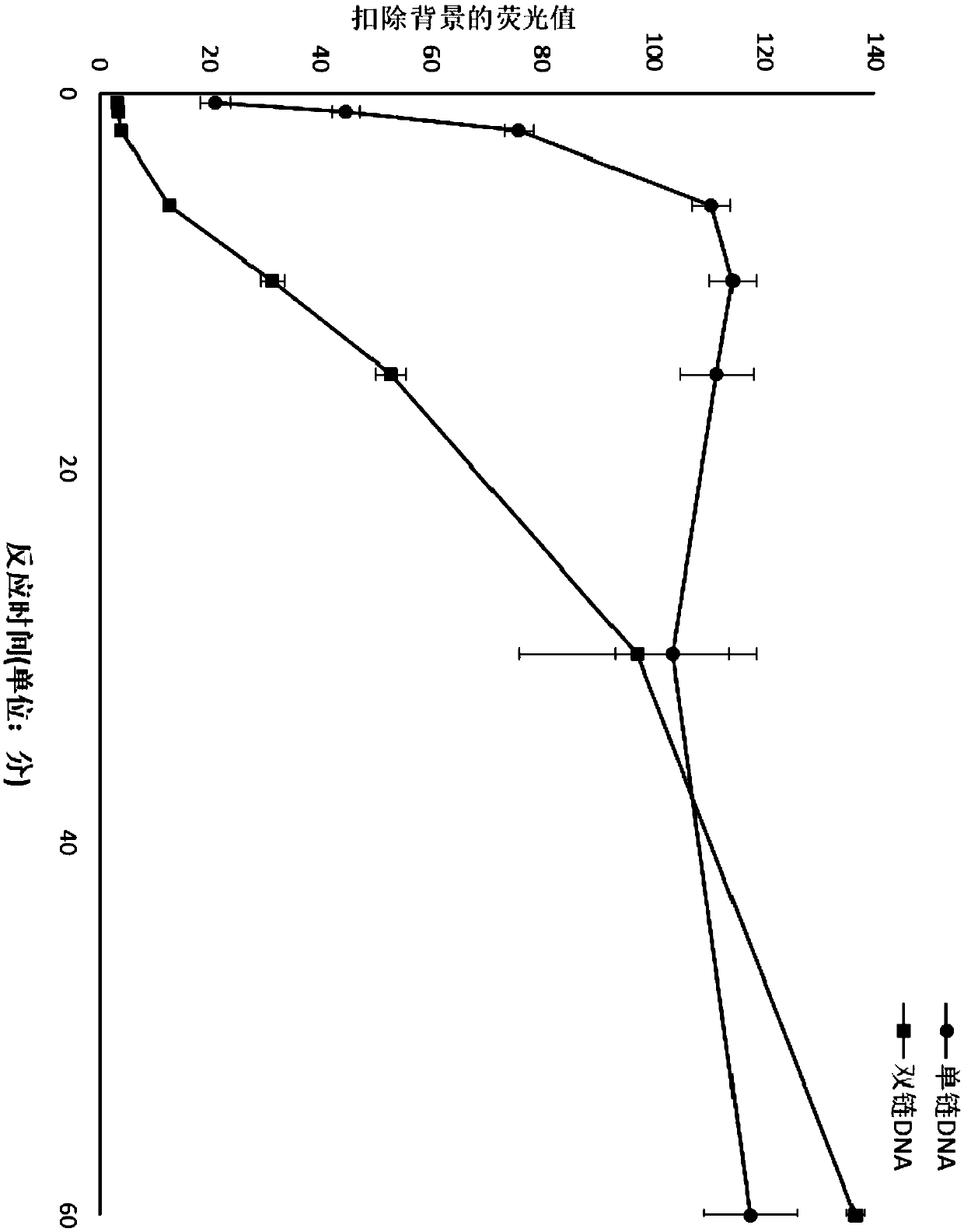
[0310] Example 2 HOLMES v2.0 (LAMP combined with Cas12b) test response sensitivity
[0311] By detecting the excited fluorescence intensity of the fluorescent probe (HEX-N12-BHQ1), determine the target DNA concentration required for Cas12b to exercise bypass single-stranded DNA cleavage activity, that is, the sensitivity of Cas12b bypass single-stranded DNA cleavage reaction.
[0312] 1. Preparation of sgRNA
[0313] First, using the previously constructed plasmid pUC18-sgRNA-T1 as a template, primers named sgRNA-DNMT1-3-F and sgRNA-DNMT1-3-R were designed, and 20% of the target DNA in the sgRNA targeting T1 was PCR-generated. 1 base was replaced by sgRNA targeting DNMT1-3, and another plasmid pUC18-sgRNA-DNMT1-3 was obtained.
[0314] Then, using the plasmid pUC18-sgRNA-DNMT1-3 as a template, and T7-crRNA-F and ZLsgRNA-DNMT1-3-R as primers, the DNA template required for in vitro transcription was amplified by PCR. Then add DpnI to the PCR product (1 μl DpnI (10U / μl) per 50 ...
Embodiment 3
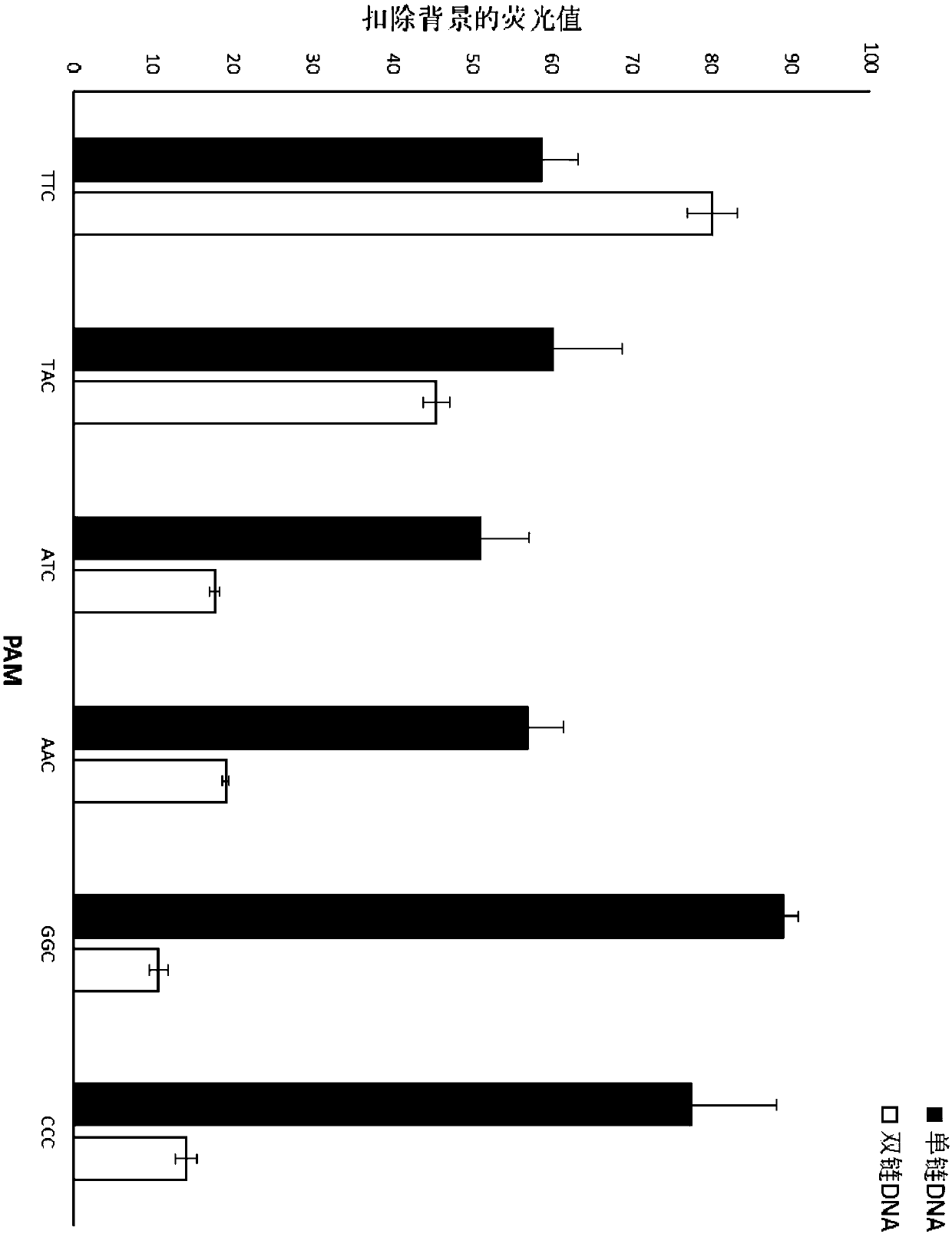
[0332] Embodiment 3Cas12b test single base mutation target
[0333] Select single-stranded DNA or double-stranded DNA as the target sequence, and test the change of Cas12btrans cutting fluorescence signal when there is a single-base mutation in the target DNA, so as to detect the single-base mutation.
[0334] 1. Preparation of guide RNA (sgRNA)
[0335]Using the plasmid pUC18-sgRNA-DNMT1-3 as a template, T7-crRNA-F as an upstream primer, and oligonucleotides containing guide sequences complementary to different targets as a downstream primer, the DNA required for in vitro transcription was amplified by PCR template. Then add DpnI to the PCR product (1 μl DpnI (10U / μl) per 50 μl PCR system), bathe in water at 37°C for 30 min, digest the plasmid DNA template, and recover the PCR product using the Gel and PCR Product Column Recovery Kit (Promega). product. Using this PCR recovery product as a template, use T7 High Yield Transcription Kit (Thermo) to synthesize full-length sgR...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com