Construction method and application of bacterial promoter reporter vector
A construction method and promoter technology, applied in the biological field, can solve the problems of unstable fluorescent quantitative PCR results, expensive fluorescent dyes, and high cost, and achieve stable and reliable results, low cost, and strong inclusiveness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment 1, the construction of reporter carrier
[0033] S1. Construction of reporter vector pBBRMCS-lacZ
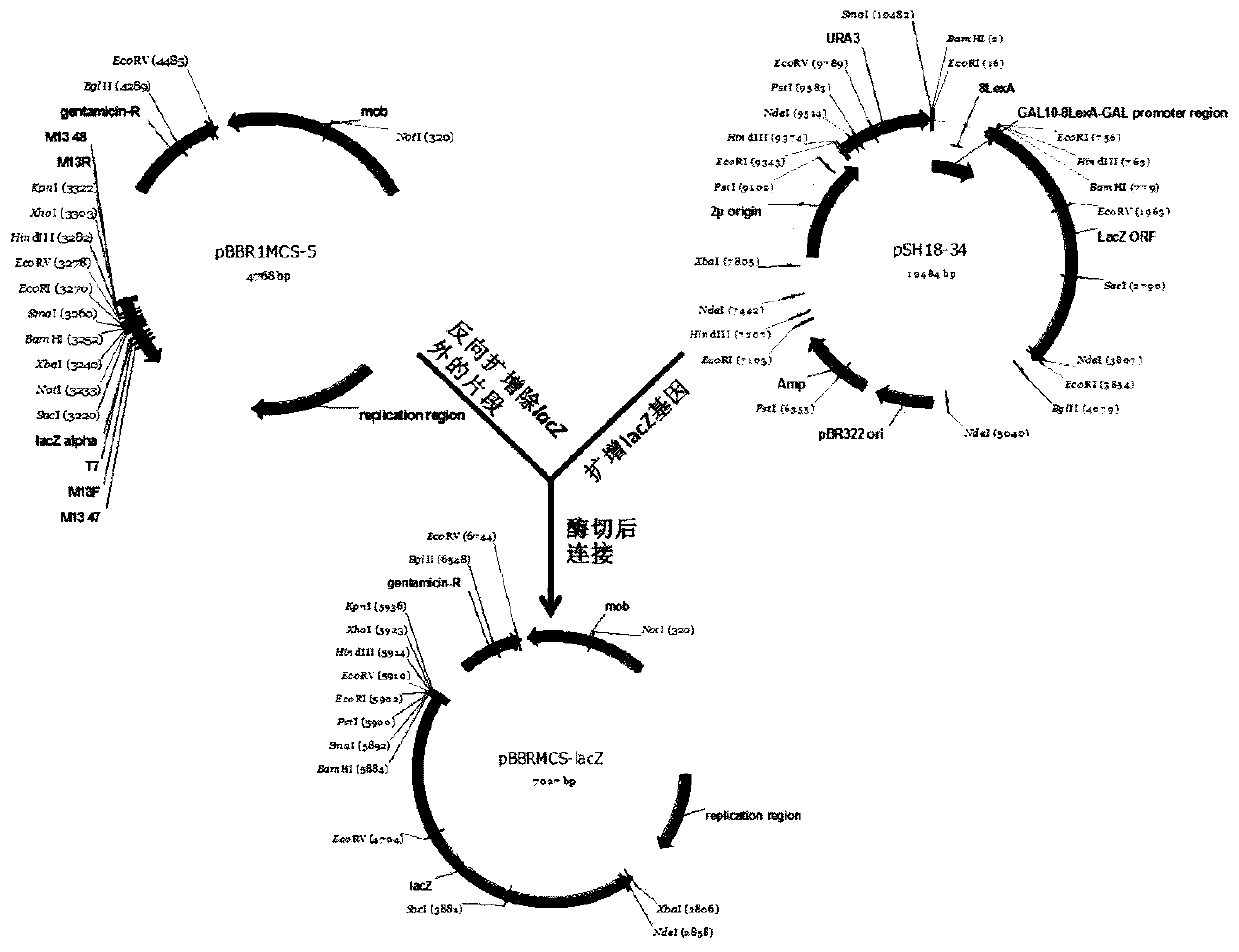
[0034] The schematic diagram of the construction of the reporter vector pBBRMCS-lacZ is as follows: figure 1 As shown, using the primer pair P1 / P2, using the pBBR1MCS-5 plasmid as a template, the plasmid backbone fragment was obtained by reverse amplification, including the gentamicin resistance gene, replicon and mob transfer gene; using the primer pair P3 / P4, The lacZ gene was amplified using the pSH18-34 plasmid as a template; the above two PCR products were digested with KpnI and XbaI, ligated with T4 DNA ligase, and verified by sequencing to obtain the vector pBBRMCS-lacZ. Primer P3 has recognition sequences of 7 common restriction endonucleases (KpnI, XhoI, HindIII, EcoRI, PstI, SmaI and BamHI), which provide multiple cloning sites for the vector after ligation. Such as figure 1 In the reporter vector pBBRMCS-lacZ shown, the tac promoter and lacI const...
Embodiment 2
[0049] Embodiment 2, detecting the promoter activity of fleS gene
[0050] S1. Construction of the reporter vector pBBRMCS-fleSpro-lacZ
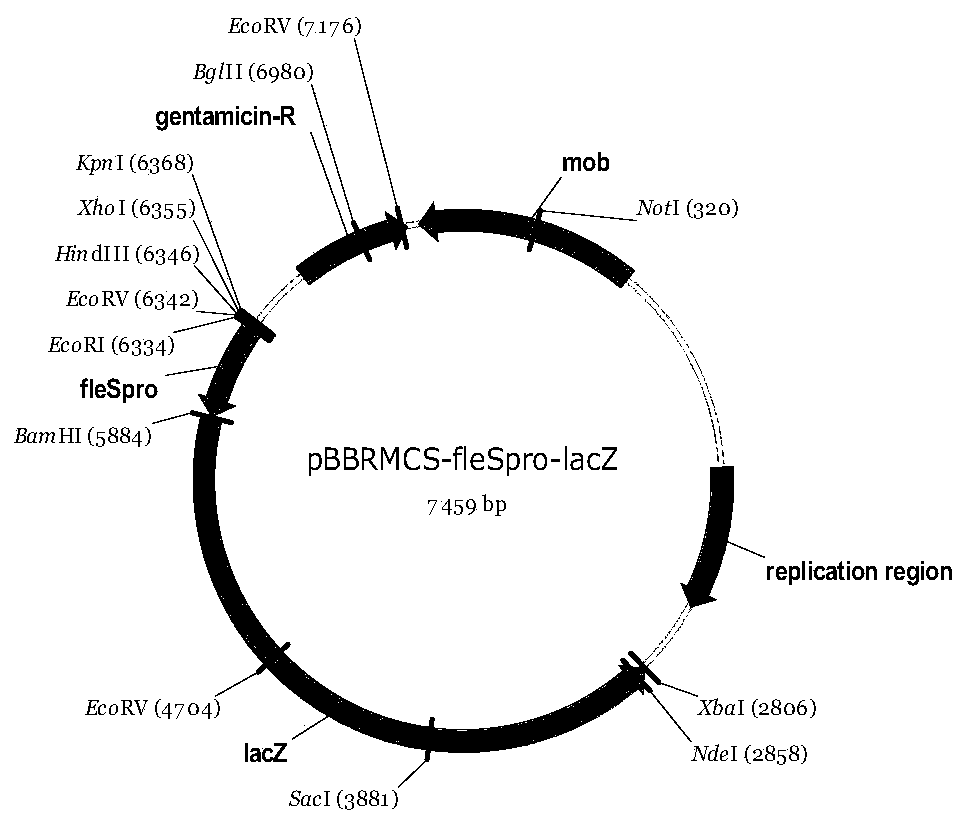
[0051] The map of the reporter vector pBBRMCS-fleSpro-lacZ is as follows image 3 shown. FleS is a histidine kinase in Pseudomonas, and its gene promoter activity is positively regulated by FleQ, and the loss of fleQ will lead to a decrease in fleS promoter activity. The present invention uses the fleS promoter as the target promoter to test the feasibility of the present invention. Using the total genomic DNA of Pseudomonas putida KT2440 as a template, the promoter of the fleS gene was amplified using the primer pair P5 / P6, and then ligated into the pBBRMCS-lacZ vector with BamHI and EcoRI, and sequenced to verify that the promoter sequence was correct. The reporter vector pBBRMCS-fleSpro-lacZ was obtained.
[0052] S2. Detecting the activity of the fleS promoter
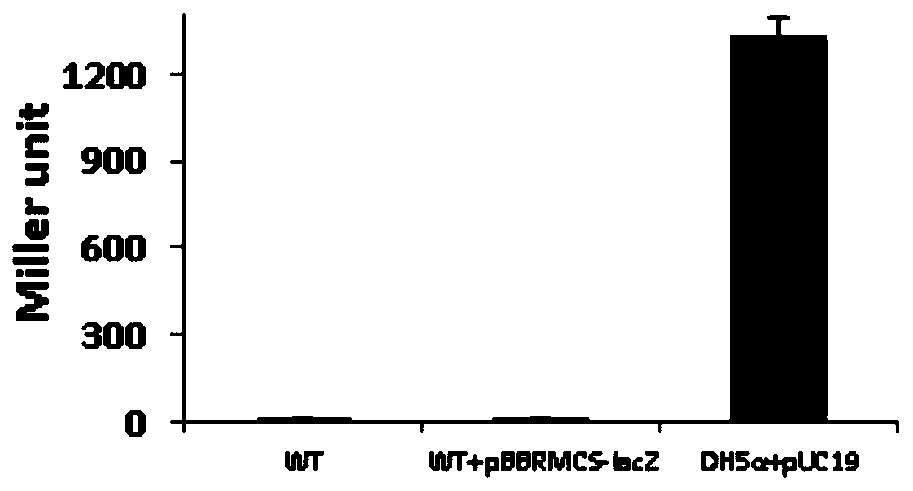
[0053] The pBBRMCS-fleSpro-lacZ vector was introduced into the wild-typ...
Embodiment 3
[0054] Example 3, detection of whether the unknown DNA fragment has promoter activity
[0055] S1. Construction of reporter vectors pBBRMCS-PP_1493pro-lacZ and pBBRMCS-PP_1494pro-lacZ
[0056] PP_1493 and PP_1494 are two genes in the wsp operon of Pseudomonas putida KT2240. PP_1494 is responsible for the synthesis of the second messenger c-di-GMP, and PP_1493 inhibits the synthesis activity of PP_1494. The two genes are closely related to the formation of biofilm . Whether the sequences in front of the PP_1493 and PP_1494 genes have promoter activity is still unclear. The present invention detects whether an unknown sequence upstream of the PP_1493 and PP_1494 genes has promoter activity to test the feasibility of the present invention. Use the primer pair P7 / P8 to amplify the upstream 500bp sequence of the PP_1493 gene, digest it with EcoRI+BamHI, and connect it to the pBBRMCS-lacZ vector to obtain pBBRMCS-PP_1493pro-lacZ. The plasmid map is as follows Figure 5 shown. Use...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com