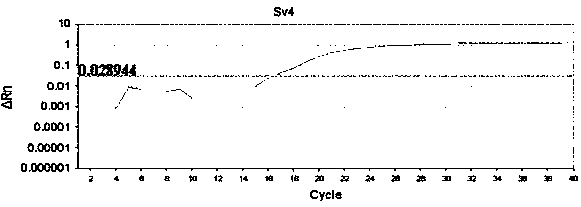
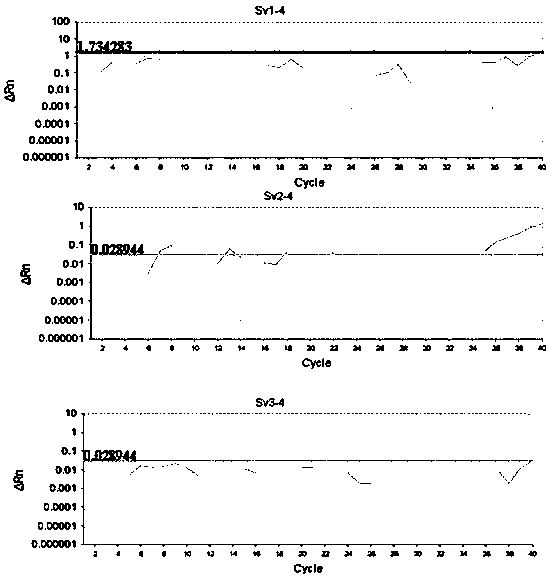
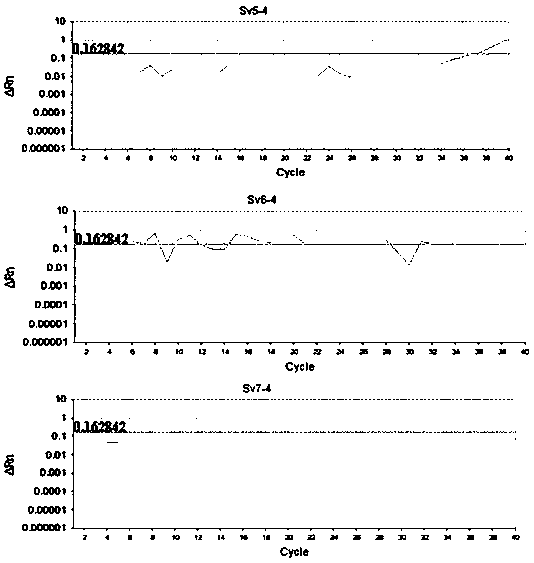
Detection method formolecular typing of acinetobacter baumannii (Ab) Sv4-serotype O antigen
A technology for Acinetobacter baumannii, serotypes, applied in microorganism-based methods, biochemical equipment and methods, and microbial assay/inspection, etc. Highly repeatable and accurate results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Design of primers and probes
[0026] 1. Screening of specific genes
[0027]Acinetobacter baumannii Sv4 serotype O antigen synthesizes O antigen through two pathways, one is wzy The O antigen is synthesized through the ABC-transporter pathway, and the O antigen is synthesized through the ABC-transporter pathway. In the former, wxya / wzy Genes are very specific and can be directly used as candidate genes for designing probes, and in the latter, because there is no wxya / wzy Such a specific gene, so only relatively specific genes can be selected, usually some rare monosaccharide genes or glycosyltransferases. In the present invention, the all_vs_all_blast method is performed on all the genes in the gene cluster for comparison, and the matching number of the specific gene must be much smaller than that of the conservative gene. Combine the above methods to find specific genes and design probe primers for them.
[0028] 2. Design of primers and probes
[00...
Embodiment 2
[0037] Extraction of sample nucleic acid (separated from any environment suitable for the life of Acinetobacter baumannii, the crude extract of the pure culture of the sample)
[0038] 1. Sample processing: Take 1 mL of the overnight cultured bacterial solution and add it to a 1.5 mL centrifuge tube, centrifuge at 8000 rpm for 1 min at room temperature, discard the supernatant, and collect the bacteria. Add 400 µL Buffer Digestion, vortex to mix, and bathe in water at 65 °C for 1 h until the cells are completely lysed.
[0039] During the water bath process, invert and mix once every 10 minutes, which can promote the lysis of the sample, and the mixed solution becomes clear and transparent, indicating that the lysis is complete;
[0040] 2. Add 200 µL Buffer PB, mix thoroughly by inversion, and place in -20 ℃ refrigerator for 5 min;
[0041] 3. Centrifuge at 10,000 rpm for 5 minutes at room temperature, and transfer the supernatant (500-550 µL) to a new 1.5 mL centrifuge tube...
Embodiment 3
[0049] Sample PCR amplification
[0050] Use the extracted nucleic acid solution as a template for PCR reaction, PCR reaction system and reaction conditions:
[0051] PCR reaction system (25 µL):
[0052]
[0053]
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com