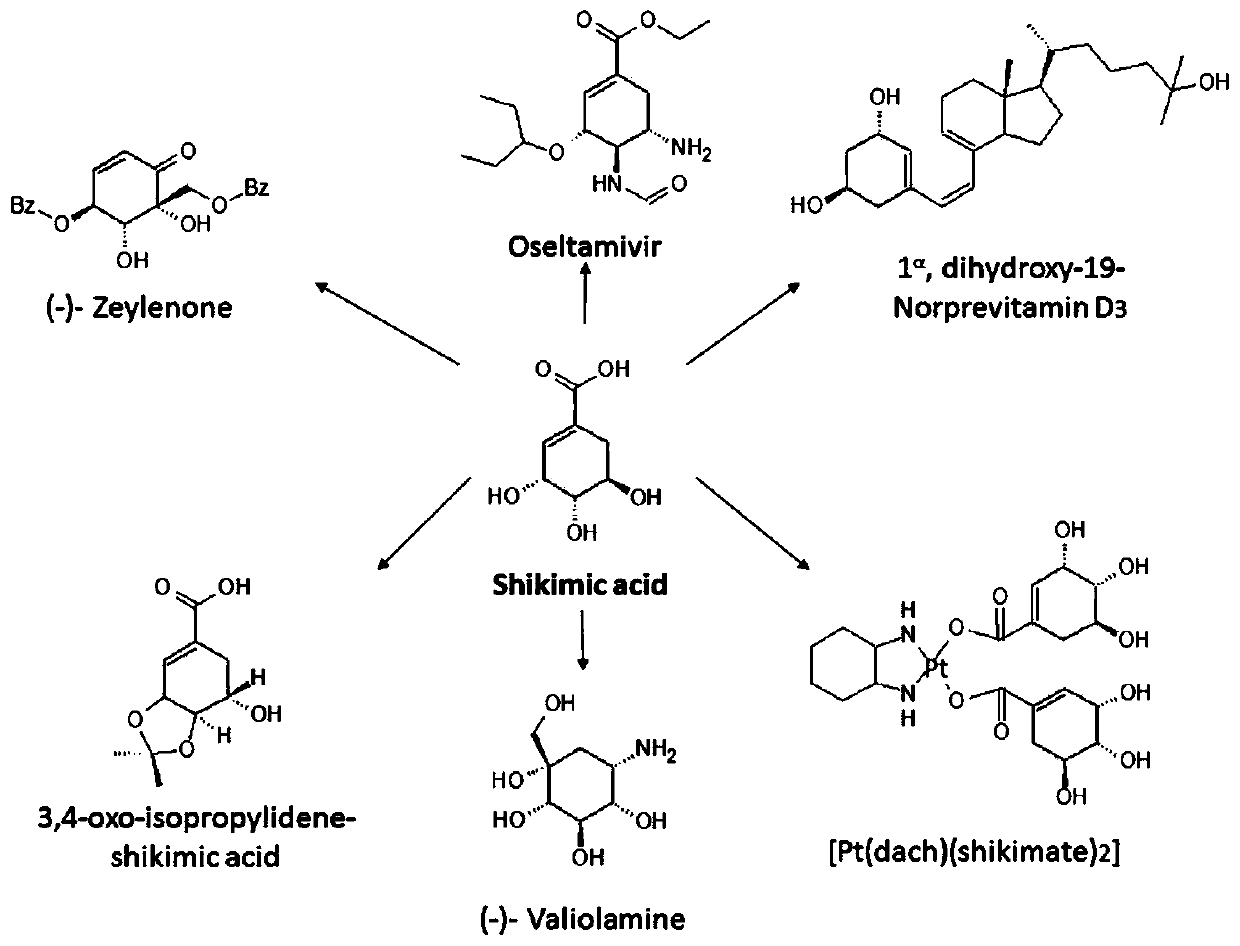
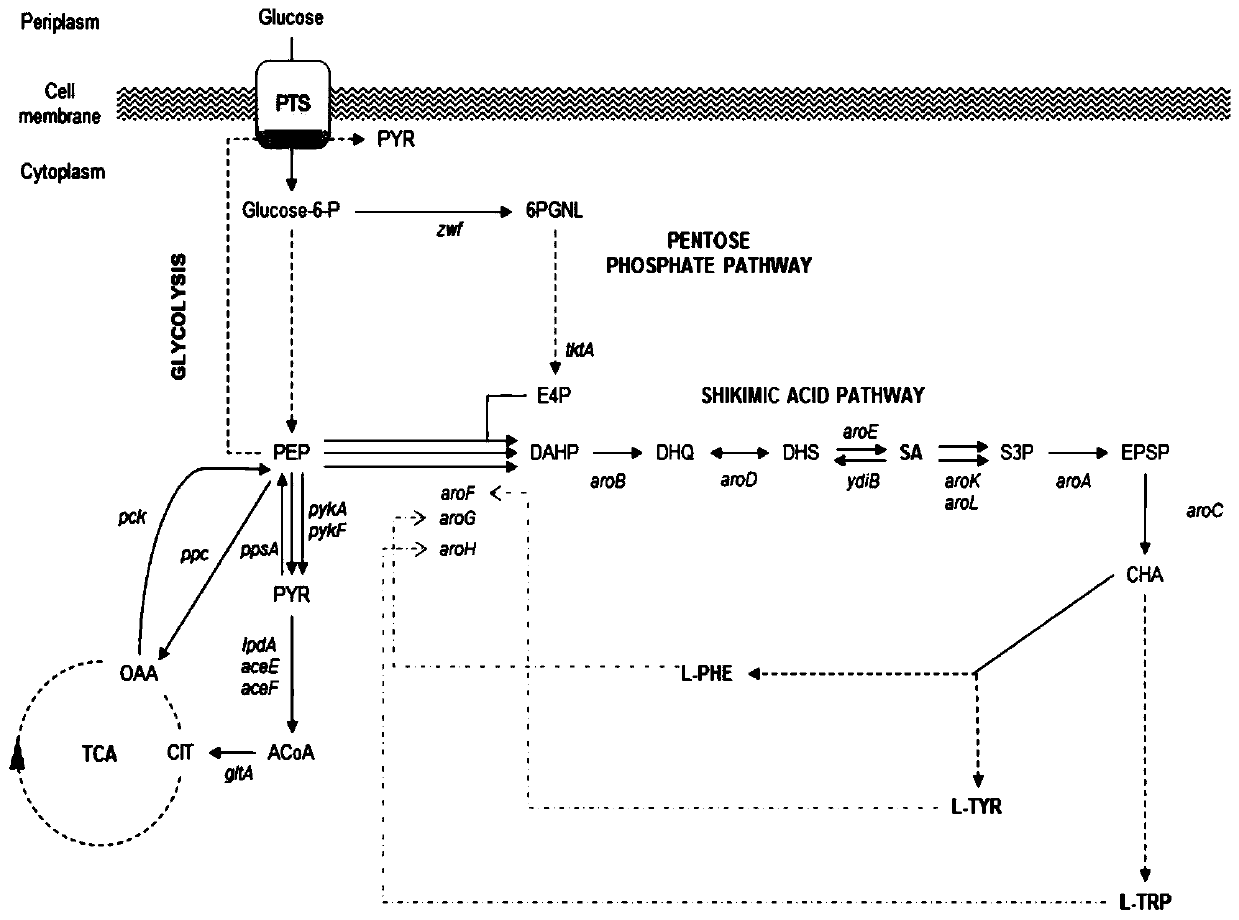
Recombinant strain and application thereof
A technology for recombining strains and strains, applied in the field of bioengineering, can solve problems such as unfavorable by-products, unsatisfactory yield, etc., and achieve the effects of increasing fermentation yield, improving survival rate and cell state, and increasing yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0104] Example 1 Construction of Escherichia coli CRISPR-Cas9 system
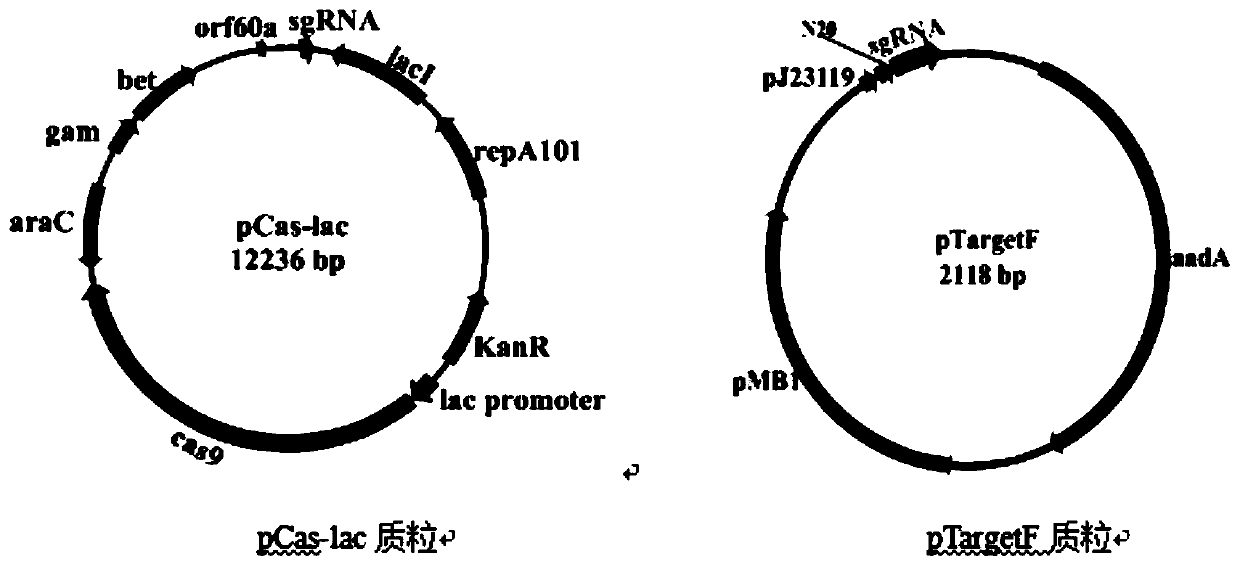
[0105] The Escherichia coli CRISPR-Cas9 system consists of two basic plasmids, pCas-lac and pTargetF, which were constructed according to the method in the literature "Multigene Editing in the Escherichia coli Genomeviathe CRISPR-Cas9 System". Plasmid pCas-lac is an episomal plasmid of Escherichia coli, which contains L-arabinose-induced expression of Red recombinase element, Cas9 protein coding gene Cas9, temperature-sensitive element, sgRNA used for induction and elimination of plasmid pTargetF, and kanamycin resistance Gene KanR et al. All plasmid series containing pCas-lac derivatives need to be cultured at 30°C to ensure normal replication of the plasmid without loss.
[0106] Plasmid pTargetF is an Escherichia coli episomal plasmid, containing the spectinomycin resistance gene aadA and the promoter pJ23119 for transcription of sgRNA, see the plasmid map image 3 .
[0107] According to the sequence...
Embodiment 2
[0129] Example 2 Construction of Escherichia coli BL21ΔaroL (ESA-1) Engineering Bacteria
[0130] Primers were designed according to the upstream and downstream sequences of Escherichia coli BL21(DE3) aroL gene. According to the genome sequence of Escherichia coli BL21(DE3) published on NCBI, the sequence of shikimate kinase aroL was found, the cleavage site N20 was selected on the aroL gene, and the upstream homology arm and downstream homology arm of aroL knockout were amplified by PCR , the primers are as follows:
[0131] Amplification of aroL upstream homology arm primers
[0132] SA-001: GGTATAGTAAGGGGTGTATTGAG
[0133] SA-002: ACAAATAAACCACGATCCCGAGGGCCATTCCGACCGTTGT
[0134] Primers used to amplify the downstream homology arm of aroL
[0135] SA-003-aroL: ACAACGGTCGGAATGGCCCTCGGGATCGTGGTTTATTTGT
[0136] SA-004-aroL: GACGCAGATCCCTTGTAGTG
[0137] Amplification of DonorDNA
[0138]
[0139] Primer annealing yields the cleavage site N20-sequence:
[0140] SA-0...
Embodiment 3
[0144] Example 3 Construction of Escherichia coli BL21ΔptsHIcrrΔaroL (ESA-2) Engineering Bacteria
[0145] Primers were designed according to the upstream and downstream sequences of Escherichia coli BL21(DE3) ptsHIcrr gene. According to the genome sequence of Escherichia coli BL21(DE3) published on NCBI, the glucose utilization gene cluster ptsHIcrr sequence was found, the cutting site N20 was selected on the crr gene, and the upstream homology arm and downstream homology arm of ptsHIcrr knockout were amplified by PCR Arm (homologous arm includes the upstream and downstream of the entire ptsHIcrr gene cluster, after homologous recombination, the ptsHIcrr gene cluster can be knocked out), the primers are as follows:
[0146] SA-18: GTGCCGTCTACTGGCGCGACATTGTATTTCCCCAACTTATAGG
[0147] SA-21: GAGATATGGGAAGATACCGACG
[0148] SA-19::CCTATAAGTTGGGGAAATACAATGTCGCGCCAGTAGACGGCAC
[0149] SA-22: CCAGCAGCATGAGAGCGATG
[0150] Primer annealing yields the cleavage site N20-sequence: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com