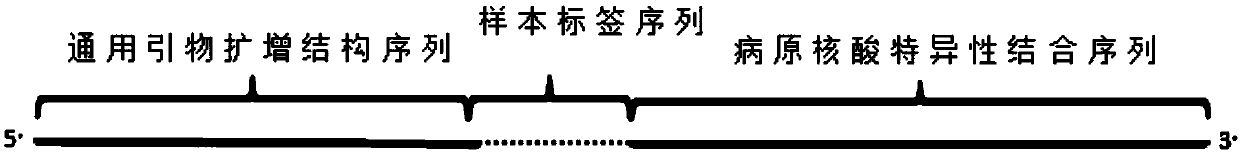
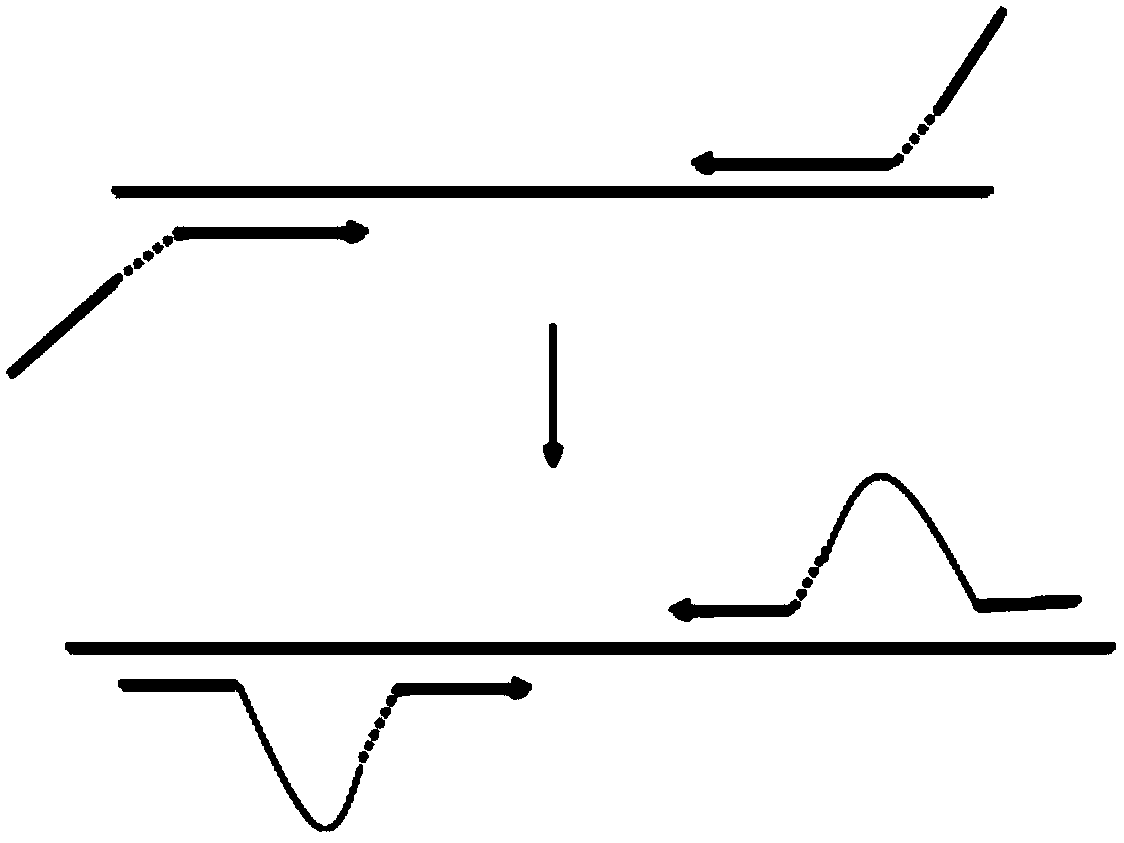
Primer group for amplifying pathogenic nucleic acid, construction method of pathogenic nucleic acid detection library and pathogenic detection method
A technology for amplifying primers and primer sets, which is applied in the field of pathogen detection, can solve problems such as expensive detection costs, complex operations, and insufficient detection throughput, and achieve the effects of reducing detection costs, simplifying technical processes, and improving detection throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Embodiment 1: the typing detection of pathogenic microorganism (HPV nucleic acid typing detection)
[0076] 1. Sample Preparation
[0077] Prepare 1000, 100, and 10 copies / microliter of national standard products for the L1 region of human papillomavirus nucleic acid, including HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, 6, 11, 26, 53, 73, 82, and prepare 30 HPV clinical DNA samples with known HPV typing test results at the same time. A total of 90 cases of national standard products and HPV clinical DNA samples with different gradient concentrations.
[0078] 2. Multiplex PCR amplification
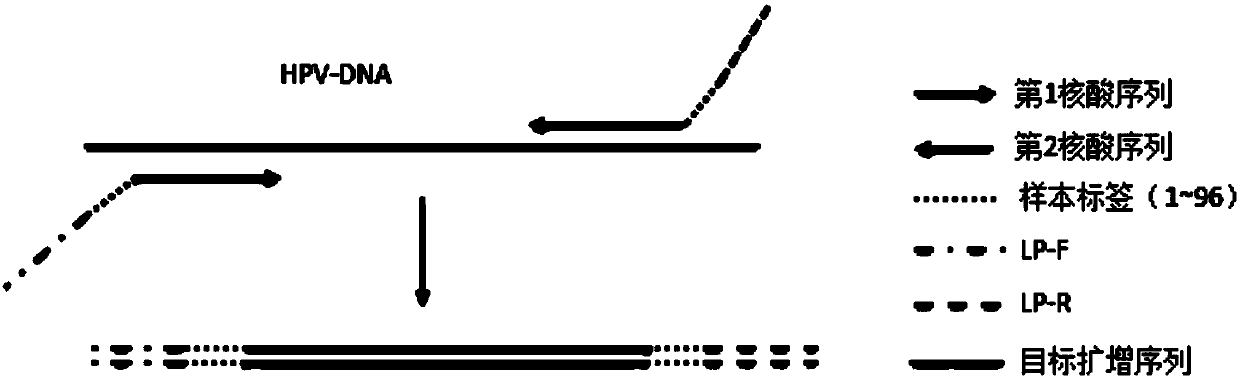
[0079] The prepared 90 samples and 6 blank controls were numbered sequentially, and 96 sets of forward primers and reverse primers with sample tag sequences were respectively amplified to obtain 96 multiplex PCR amplification products, such as image 3 shown.
[0080] It should be noted that different sample tag sequences are used for different DNA samples, and t...
Embodiment 2
[0117] Embodiment 2: the identification of pathogenic microorganism (HCV detects)
[0118] 1. Sample Preparation
[0119] Use the extraction reagent of the hepatitis C virus nucleic acid assay kit (PCR-fluorescent probe method) of Sun Yat-sen University Daan Gene Co., Ltd. to extract, according to the instructions provided by the manufacturer, from 48 cases of serum with known HCV detection results Template RNA was extracted from samples, and HCV clinical serum samples were provided by Tianjin Huada Clinical Inspection Center Co., Ltd.
[0120] 2. Reverse transcription reaction
[0121] The prepared 48 samples were numbered sequentially, and the cDNA of the samples was obtained by using the MMLV reverse transcriptase of Treasure Bioengineering (Dalian) Co., Ltd. and adding random primers (N8, see Table 13) for reverse transcription reaction.
[0122] The reverse transcription reaction system is 20 microliters, and its composition is shown in Table 11 below:
[0123] Table 1...
Embodiment 3
[0160] Embodiment 3: Bacterial drug resistance gene monitoring (Escherichia coli drug resistance gene detection)
[0161] 1. Sample Preparation
[0162] This implementation case uses mutant E. coli strains (ATCC43888) and wild-type E. coli strains (ATCC8739) from the China Food and Drug Control Institute, wherein the mutant E. coli strains include caiC807, flhA1281, hybA612, valS528, valS1317, valS1356, valS1368 , valS1389, yafE227, and yedK1323, a total of 10 mutation sites, were extracted using the nucleic acid purification kit produced by Huada Biotechnology (Wuhan) Co., Ltd., and operated in strict accordance with the instructions provided by the manufacturer. Use Qubit3.0 to measure the concentration of DNA samples, and then use TE buffer to dilute to 7000 and 5000 copies / microliter respectively. Then the mutant E. coli strain (ATCC43888) and the wild-type E. coli strain (ATCC8739) at the same concentration were respectively 1:0, 3:1, 1:1, 1:3, 1:9, 1:20, 1: 99. Mix at ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com