Cellulase gene gk2691 for encoding cellulase family GH30, and application thereof
A technology of gk2691 and GK2691, which is applied to the cellulase gene gk2691 encoding the cellulase family GH30 and its application fields, can solve the problems of low activity and low glycosidase content, and achieve simple material selection, effective utilization, and improved degradation efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1: Cellulase gene gk2691 the acquisition of
[0024] (1) Weigh 25g of living coral samples, place them in a 50mL sterile centrifuge tube, add 20mL of sterile seawater, and vortex for 5-10min to completely disperse the coral tissue in the seawater; then dilute the shaken seawater 100 times Afterwards, 2216E plates were coated and cultured at room temperature for 24 hours to grow single colonies;
[0025] (2) Screen a single colony on the aescin plate, and the colony that makes the medium turn black is the functional colony; select the functional colony to purify on the 2216E plate, and transfer the functional colony to the MA medium after purification Continue to purify, and finally use a DNA kit to extract the DNA of the functional bacteria, and perform high-throughput sequencing to obtain the cellulase gene gk2691 ;
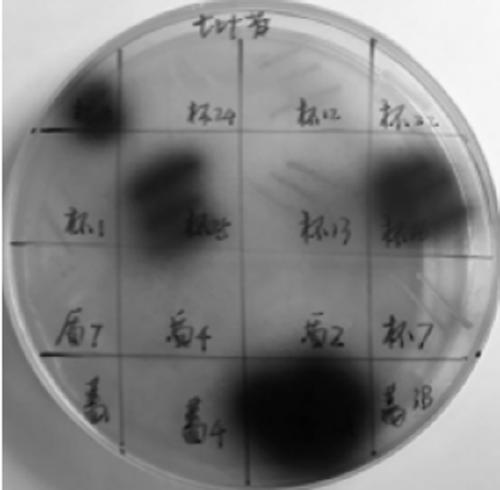
[0026] Such as figure 1 Shown is the function screening diagram of the aescin plate. Single colonies were screened out from a single strain ...
Embodiment 2
[0027] Example 2: Cellulase gene gk2691 cloning and expression.
[0028] The genes obtained through high-throughput sequencing were compared by NCBI to obtain the cellulase amino acid sequence (SEQID NO.2). If the amino acid sequence was correct, it was the target gene, and then primers were designed for it.
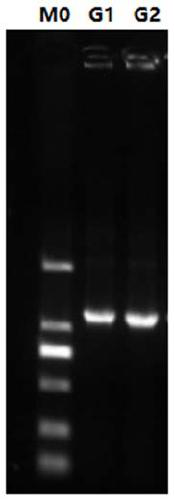
[0029] with upstream primer gk2691 F: GGCGGAATTATGAAAATGACCTCTCTCATGGCGTTATC, downstream primer gk2691 R: GCCGAAGCTTAGGGTGGCTAATTACAACTGTCTGTAACG, using high-throughput sequencing gene as template PCR amplification of cellulase gene gk2691 , followed by restriction endonuclease EcoR The amplified product was digested with Ⅰ and hind enzymes, ligated with the pET-22b expression vector digested with the same endonuclease, and then the ligated product was transformed into Escherichia coli DH5α, and spread to LB containing 50 μg / mL kanamycin Positive clones were screened on the culture medium plate, and the plasmid DNA was further extracted, and the recombinant plasmid ...
Embodiment 3
[0054] Example 3: Preparation of cellulase GK2691.
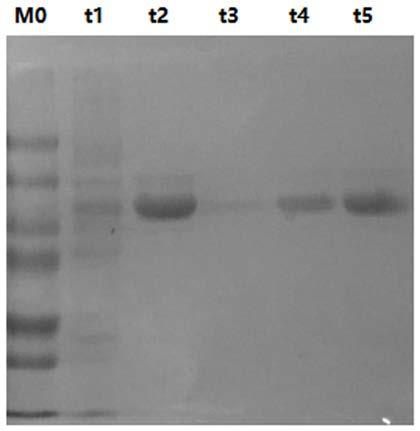
[0055] After 12 hours of incubation, the induced cells were collected at 8000 r, washed three times with PBS buffer, and finally 15-20 mL of PBS buffer was added to mix the cells, and the cells were broken with an ultrasonic cell breaker in an ice bath (power: 23%, working time: 20min). Then centrifuge at 4°C and 8000r for 15min, collect the supernatant, and obtain the crude enzyme solution. Finally, the substrate was used to detect the enzyme activity of the crude enzyme. The crude enzyme liquid was purified by a nickel chloride purification column to obtain pure enzyme, which was then desalted in a currency desalting column to obtain the final product cellulase GK2691. Finally, polyacrylamide electrophoresis was used to verify the result. See image 3 .
[0056] The analysis of the effect of cellulase GK2691 on pathogenic bacteria comprises the following steps:
[0057] Collect the v545 pathogenic bacteria cultured to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com