A method for extracting early RNA of cotton fiber cell development
A cotton fiber and cell technology, applied in the field of extracting early RNA of cotton fiber cell development, can solve the problems of specificity reduction, systematic error, and limitation of transcription level at the initial stage of fiber development, etc., and achieve high RNA quality and accurate transcriptome data Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1: Cotton variety TM-1 (from Cotton Research Institute, Chinese Academy of Agricultural Sciences) Extraction and quality detection of fiber RNA at different developmental stages - 1DPA to 5DPA.
[0045] 1) Take materials. From the TM-1 plant, 40 cotton ovaries of -1DPA, 0DPA, 1DPA, 2DPA, 3DPA, 4DPA and 5DPA were respectively taken.
[0046] 2) cracking. Take seven 15mL centrifuge tubes, add 8mL of prepared lysate respectively, and mark them. For the flowers in step 1, carefully cut open the ovary wall with a blade in the ultra-clean workbench, gently clamp the fungus handle with tweezers, take out the complete ovules and put them into the corresponding centrifuge tubes.
[0047] 3) Vacuum treatment. Put the centrifuge tube in step 2 into the desiccator, vacuumize to -0.9Mpa, 10min-20min (-1DPA sample vacuumizes 10min, 0DPA sample vacuumizes 12min, 1DPA sample vacuumizes 14min, 2DPA sample vacuumizes 16min , 3DPA samples were vacuumed for 18 minutes, 4DPA and...
example 2
[0061] Example 2: Imaging observation of ovule tissue by scanning electron microscope environment
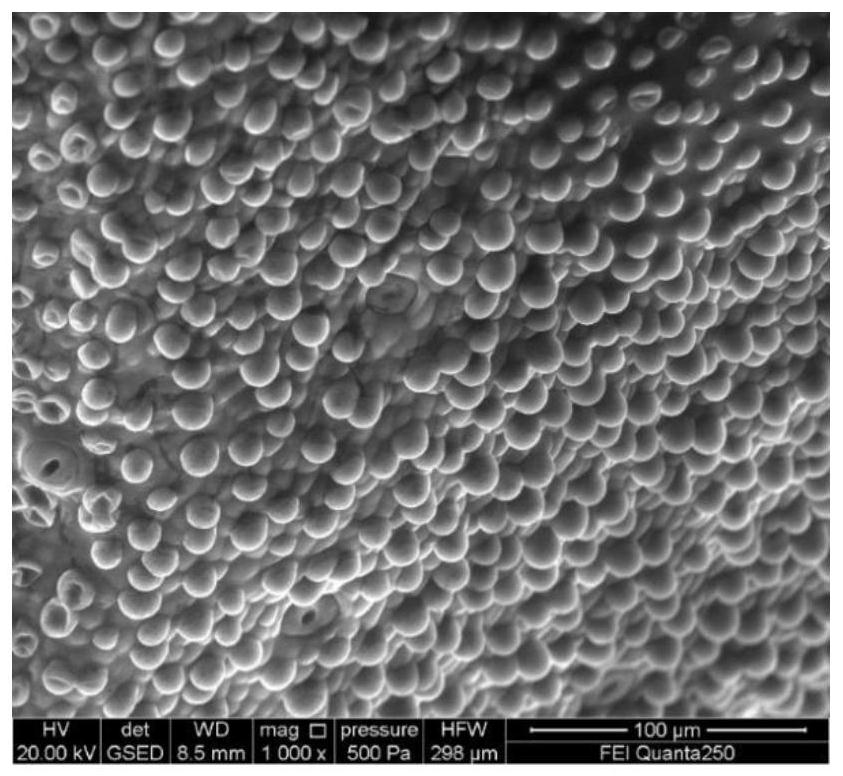
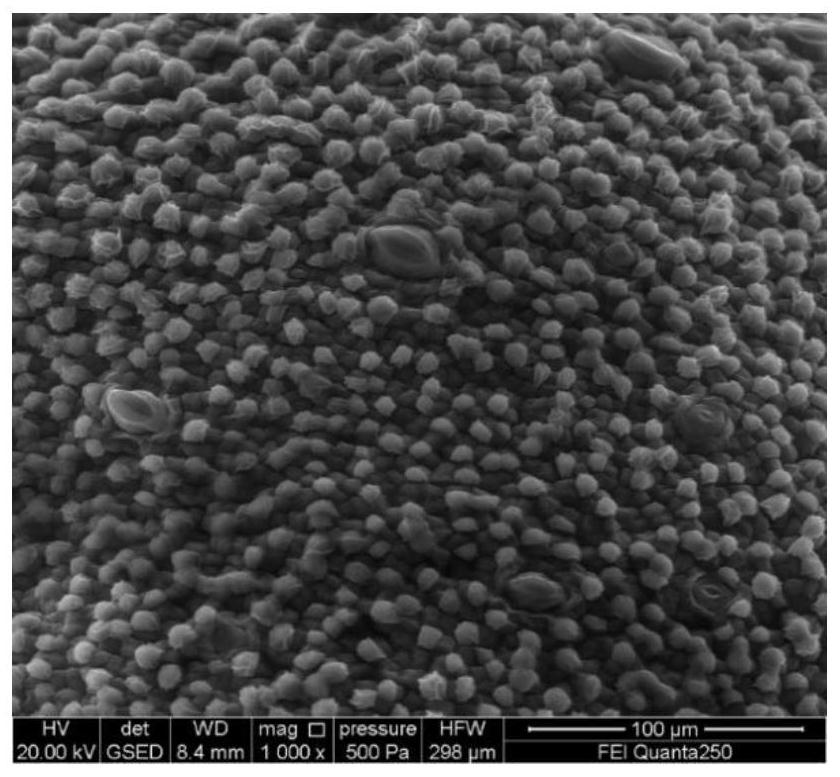
[0062] Take 1 ovary of TM-1 0DPA cotton, carefully cut open the ovary wall with a razor blade, gently clamp the fungus with tweezers, take out 6 complete ovules with the same development status, and 3 ovules are directly subjected to electron microscope environment without any treatment scanning. The other 3 ovules were lysed, put the removed ovules into a 1.5mL centrifuge tube containing 800μL lysate, vacuumed to -0.9Mpa, 12min, vortexed for 5min, ddH 2 O was washed three times, and the electron microscope environment was scanned at a magnification of 1000 times.
[0063] The results of electron microscope imaging showed that the epidermal cells of the ovules without lysis treatment had normal protrusions, and the fiber cells were evenly and plumply distributed in the epidermis ( figure 2 ). The ovule epidermis tissue after vacuum lysis treatment, because the ovule epiderma...
example 3
[0064] Example 3: Ovule semi-thin section observation
[0065] Take 1 ovary of TM-1 0DPA cotton, carefully cut open the ovary wall with a blade, gently clamp the funicular handle with tweezers, take out 6 complete ovules with the same development status, and fix the 3 ovules directly without any treatment . The other 3 ovules were lysed, put the removed ovules into a 1.5mL centrifuge tube containing 800μL lysate, vacuumed to -0.9Mpa, 12min, vortexed for 5min, fixed with fixative, sliced, and observed.
[0066] The process of semi-thin sectioning is as follows:
[0067] 1) Sample fixation. Put the ovules into a 2mL centrifuge tube filled with 1mL of fixative solution (the components of the fixative solution are 4% paraformaldehyde and 5% glutaraldehyde), and vacuumize to -0.9MPa until the ovules are completely immersed in the fixative solution.
[0068] 2) Wash. Wash 3 times with 0.1M Pbs (phosphate buffered saline), 15 min each time.
[0069] 3) Dehydration. Use 30% etha...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com