Novel genome editing tool (Lb2Cas12a-RVR) and construction method and application method thereof
A genome editing and gene editing technology, applied in the field of new genome editing tools and its construction, can solve the problems of low activity and low fidelity of specific gene editing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
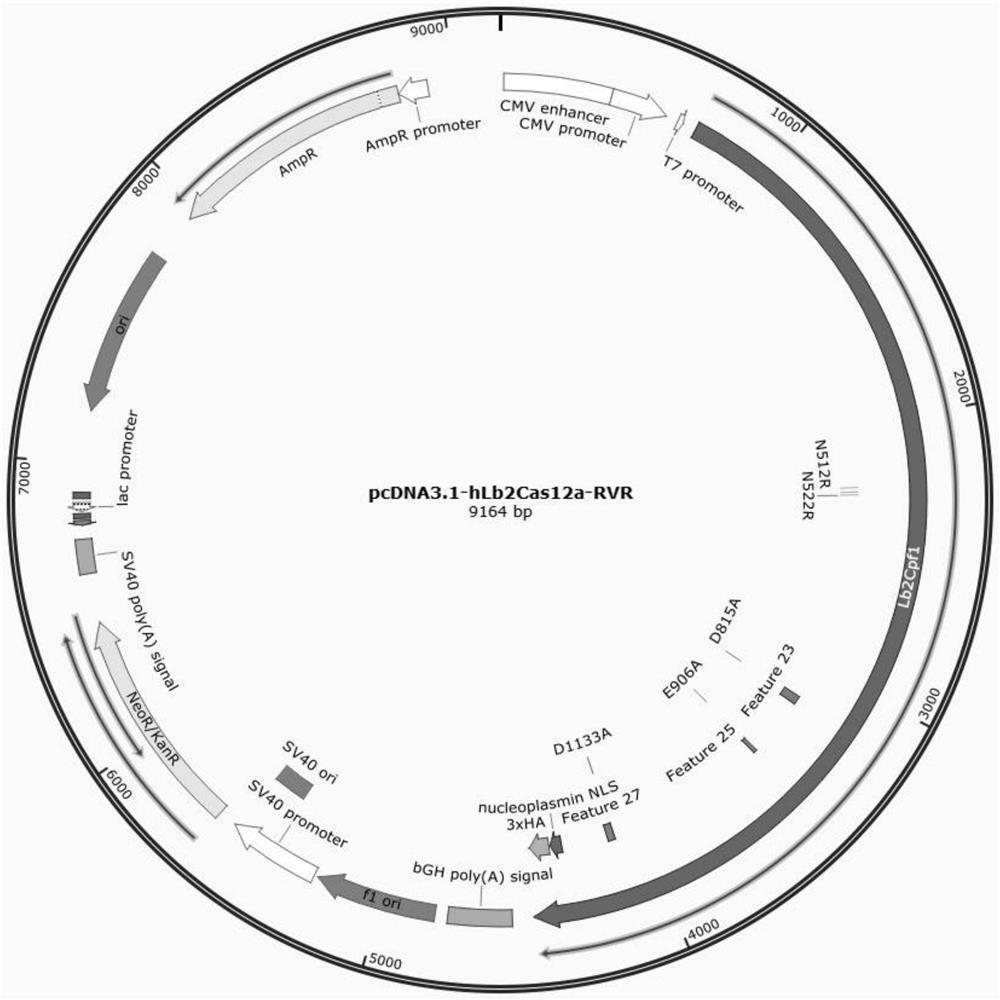
[0038] Example 1: Construction of pcDNA3.1-hLb2Cas12a-RVR plasmid
[0039] (1) Using the PCR method, set the corresponding mutation N512R+K518V+N522R on the primer, amplify the sequence containing the RVR mutation site on the pY011 (pcDNA3.1-hLb2Cas12a) vector, and pass the amplified fragment and backbone through ClonExpressII One Step Cloning Kit gets the vector pcDNA3.1-hLb2Cas12a-RVR after mutation.
[0040] (2) The PCR system used in the plasmid construction experiment is as follows:
[0041] Template: 10ng; Forward Primer: (10μM) 1μl; Reverse Primer: (10μM) 1μl;
[0042] dNTP: 0.5μl; DNA polymerase (Vazyme, P501): 0.5μl; 5x Buffer: 10μl; RNase-Water fill up to 50μl.
[0043] The PCR program is as follows:
[0044] 95℃, 3min; 95℃, 15sec, 60℃, 15sec, 72℃, 1min; 35cycles; 72℃, 3min.
[0045] The application method of a Lb2Cas12a-RVR gene editing tool of this embodiment:
[0046] Step 33: Including the construction of crRNA directed against the DNA target of a certain disease-causing gen...
Embodiment 2
[0054] Example 2: Establishment of a GFP reporter system for pcDNA3.1-hLb2Cas12a-RVR activity test
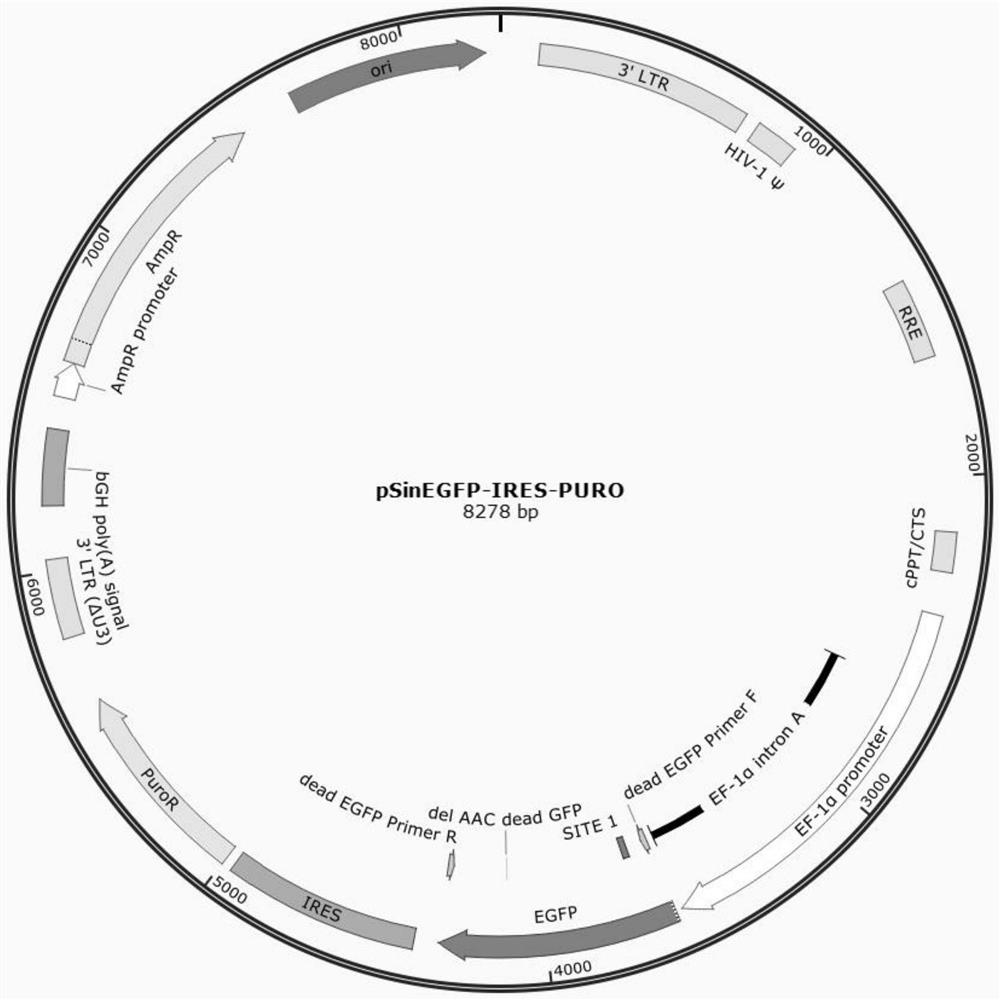
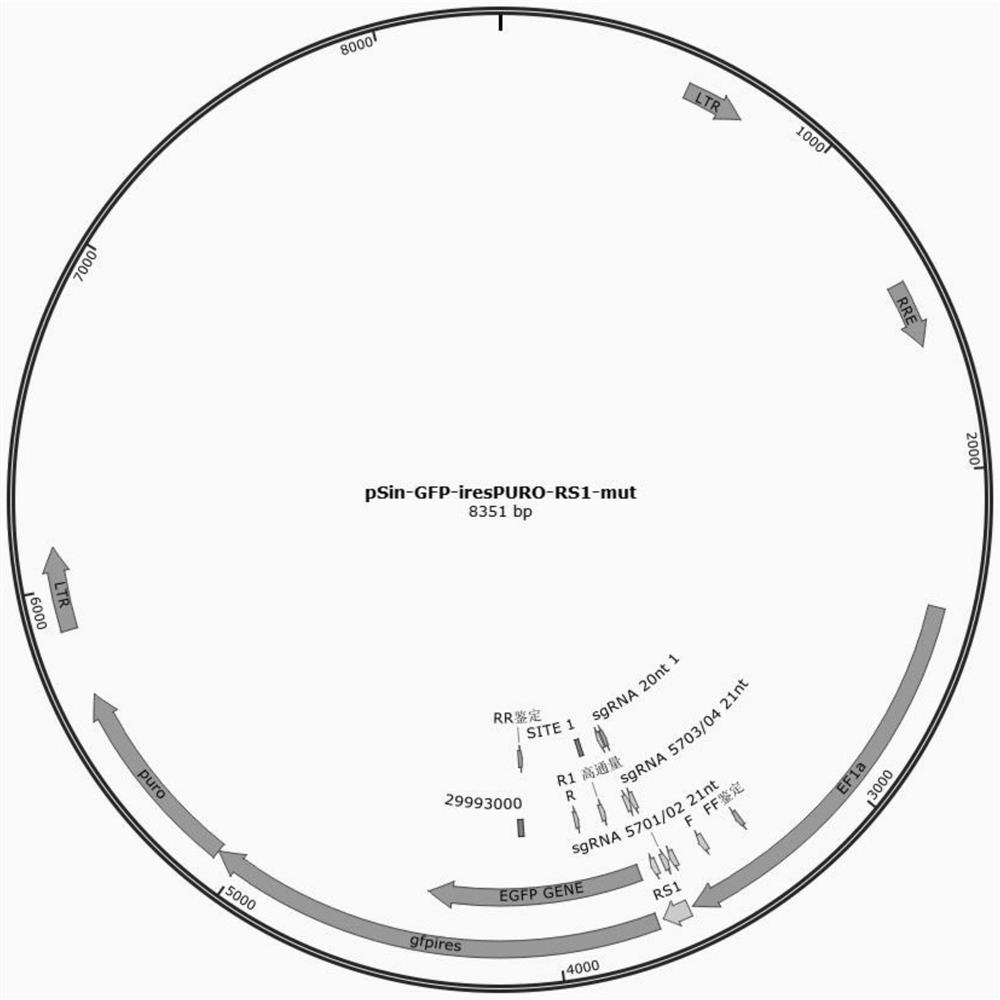
[0055] (1) Construct a pSin-GFP lentiviral vector, which contains the GFP gene (as shown in SEQ ID NO. 4), IRES and Puromycin resistance genes. pSin-GFP and the helper plasmid were co-transfected into HEK-293T cells, the virus supernatant was collected, filtered with a 0.45μm filter, and aliquoted and stored at -80°C for later use.
[0056] (2) Recovery of HEK-293 cells: Take out the tube of frozen HEK-293 cells from liquid nitrogen, immediately put it in a 37°C water bath, shake it slightly, wait until the liquid is completely melted (about 1~1.5min), take it out Wipe and disinfect with 75% alcohol and place it on the ultra-clean workbench; transfer the above cell suspension to a 15ml centrifuge tube containing 5ml culture medium, centrifuge at 1500rpm for 5min; discard the supernatant, add 1ml culture medium to resuspend the cells . Transfer to a 10cm petri dish containing 10ml ...
Embodiment 3
[0062] Example 3: Determination of the efficiency of pcDNA3.1-hLb2Cas12a-RVR at a specific site
[0063] (1) pJET-U6-sgRNA design: design crRNA at different positions of the GFP gene sequence according to the requirements of subsequent evaluation;
[0064] (2) HEK-293-SC1 cells were counted and then seeded in a 12-well plate with a cell density of 1.8×105 cells / well;
[0065] (3) After 24h of cell growth, the transfection reagent TurboFect (Thermo Fisher, #R0531) was used to transfect pcDNA3.1-hLb2Cas12a-RVR (750ng) and pJET-U6-crRNA (250ng);
[0066] (4) Eukaryotic cell transfection method: After mixing the plasmids to be transfected in proportion, mix in 50μl DMEM containing 1.5μl of transfection reagent Turbofect, pipetting and mixing, and after 15 minutes at room temperature, add 1ml DMEM containing (Gibco, C11995500CP) + 10% FBS (Gibco, 16000 044) medium, and 1.8×105 cells / well of HEK-293-SC1 cells were transfected in a 12-well plate.
[0067] (5) Change the fresh medium 24h after...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com